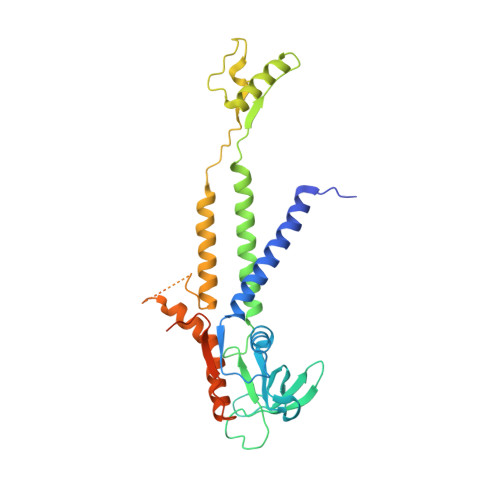
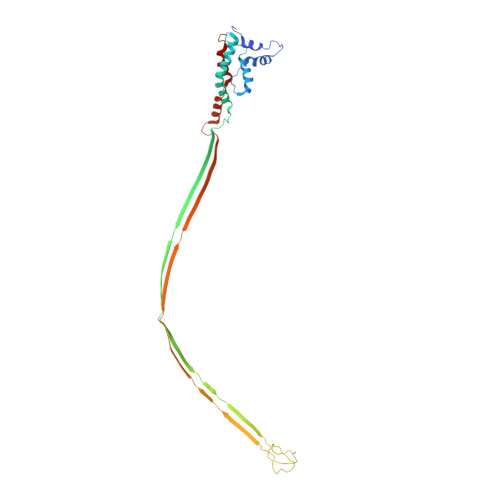
Structural assembly of the tailed bacteriophage φ29.
Xu, J., Wang, D., Gui, M., Xiang, Y.(2019) Nat Commun 10: 2366-2366
- PubMed: 31147544
- DOI: https://doi.org/10.1038/s41467-019-10272-3
- Primary Citation of Related Structures:
6QVK, 6QX7, 6QYD, 6QYJ, 6QYM, 6QYY, 6QYZ, 6QZ0, 6QZ9, 6QZF - PubMed Abstract:
The mature virion of the tailed bacteriophage ϕ29 is an ~33 MDa complex that contains more than 450 subunits of seven structural proteins assembling into a prolate head and a short non-contractile tail. Here, we report the near-atomic structures of the ϕ29 pre-genome packaging head (prohead), the mature virion and the genome-emptied virion. Structural comparisons suggest local rotation or oscillation of the head-tail connector upon DNA packaging and release. Termination of the DNA packaging occurs through pressure-dependent correlative positional and conformational changes in the connector. The funnel-shaped tail lower collar attaches the expanded narrow end of the connector and has a 180-Å long, 24-strand β barrel narrow stem tube that undergoes conformational changes upon genome release. The appendages form an interlocked assembly attaching the tail around the collar. The membrane active long loops at the distal end of the tail knob exit during the late stage of infection and form the cone-shaped tip of a largely hydrophobic helix barrel, prepared for membrane penetration.
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Center for Infectious Disease Research, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, 100084, Beijing, China.