Unravelling the receptor binding property of egg drop syndrome virus (EDSV) from the crystal structure of EDSV fiber head.
Song, Y., Wei, Q., Liu, Y., Feng, H., Chen, Y., Wang, Y., Bai, Y., Xing, G., Deng, R., Zhang, G.(2019) Int J Biol Macromol 139: 587-595
- PubMed: 31381914
- DOI: https://doi.org/10.1016/j.ijbiomac.2019.08.005
- Primary Citation of Related Structures:
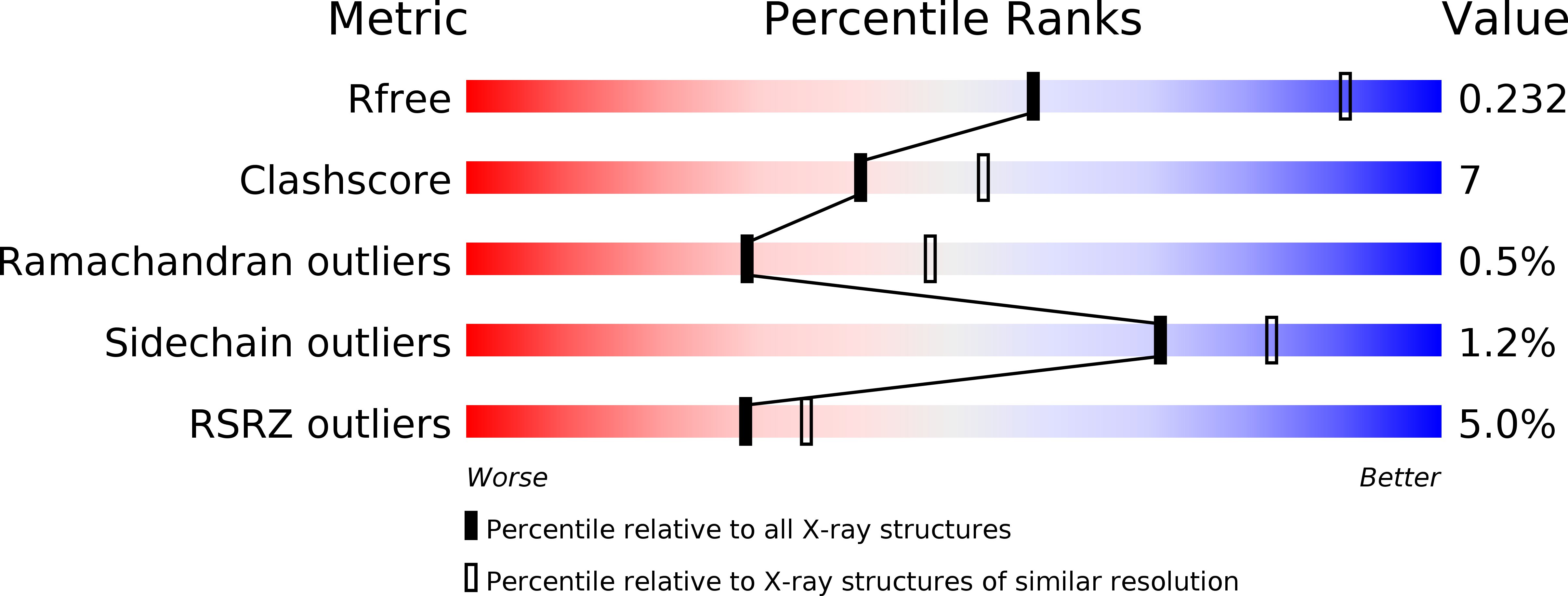
6ITX - PubMed Abstract:
Egg drop syndrome virus (EDSV) is an avian adenovirus that causes markedly decrease in egg production, and in the quality of the eggs when it infects chickens. Until now, EDSV virus-cell interactions are poorly understood, and the cellular receptor is still unknown. In the present study, we determined the atomic structure of the fiber head of EDSV (residues 377-644) at 2.74 Å resolution. Structure comparison with the (chick embryo lethal orphan) CELO long fiber head and human adenovirus fiber heads reveals that the avian adenovirus may interact with the same attachment factor in a unique fashion. Based on the previous studies of CELO virus, we assumed that the chicken coxsackievirus and adenovirus receptor (CAR) may be the attachment factor. We then demonstrate that the chicken CAR serves as a cellular attachment factor for EDSV based on three lines of evidences. Taken together, the results presented here are helpful for further exploring the pathogenesis related to the interaction between EDSV and host cells, and may be used for vaccine development and intervention strategies against EDSV infection.
Organizational Affiliation:
College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou 450002, China; Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou 450002, China.