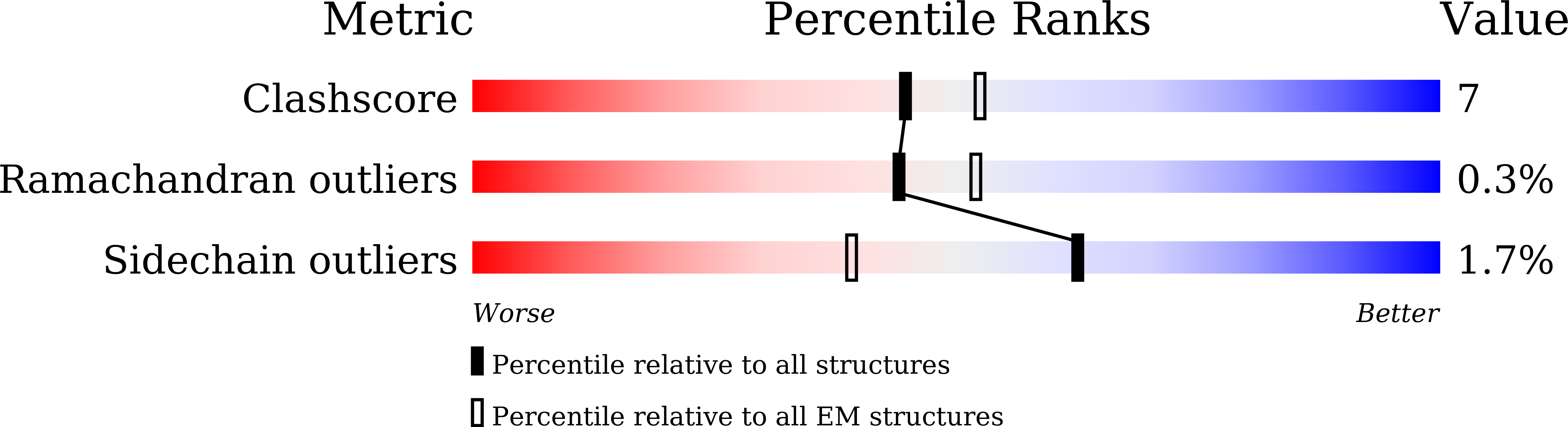Adeno-associated virus 2 bound to its cellular receptor AAVR.
Zhang, R., Cao, L., Cui, M., Sun, Z., Hu, M., Zhang, R., Stuart, W., Zhao, X., Yang, Z., Li, X., Sun, Y., Li, S., Ding, W., Lou, Z., Rao, Z.(2019) Nat Microbiol 4: 675-682
- PubMed: 30742069
- DOI: https://doi.org/10.1038/s41564-018-0356-7
- Primary Citation of Related Structures:
6IH9, 6IHB - PubMed Abstract:
Adeno-associated virus (AAV) is a leading vector for virus-based gene therapy. The receptor for AAV (AAVR; also named KIAA0319L) was recently identified, and the precise characterization of AAV-AAVR recognition is in immediate demand. Taking advantage of a particle-filtering algorithm, we report here the cryo-electron microscopy structure of the AAV2-AAVR complex at 2.8 Å resolution. This structure reveals that of the five Ig-like polycystic kidney disease (PKD) domains in AAVR, PKD2 binds directly to the spike region of the AAV2 capsid adjacent to the icosahedral three-fold axis. Residues in strands B and E, and the BC loop of AAVR PKD2 interact directly with the AAV2 capsid. The interacting residues in the AAV2 capsid are mainly in AAV-featured variable regions. Mutagenesis of the amino acids at the AAV2-AAVR interface reduces binding activity and viral infectivity. Our findings provide insights into the biology of AAV entry with high-resolution details, providing opportunities for the development of new AAV vectors for gene therapy.
Organizational Affiliation:
MOE Laboratory of Protein Science and Collaborative Innovation Center of Biotherapy, School of Medicine, Tsinghua University, Beijing, China.