Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
Kekez, M., Zanki, V., Kekez, I., Baranasic, J., Hodnik, V., Duchene, A.M., Anderluh, G., Gruic-Sovulj, I., Matkovic-Calogovic, D., Weygand-Durasevic, I., Rokov-Plavec, J.(2019) FEBS J 286: 536-554
- PubMed: 30570212
- DOI: https://doi.org/10.1111/febs.14735
- Primary Citation of Related Structures:
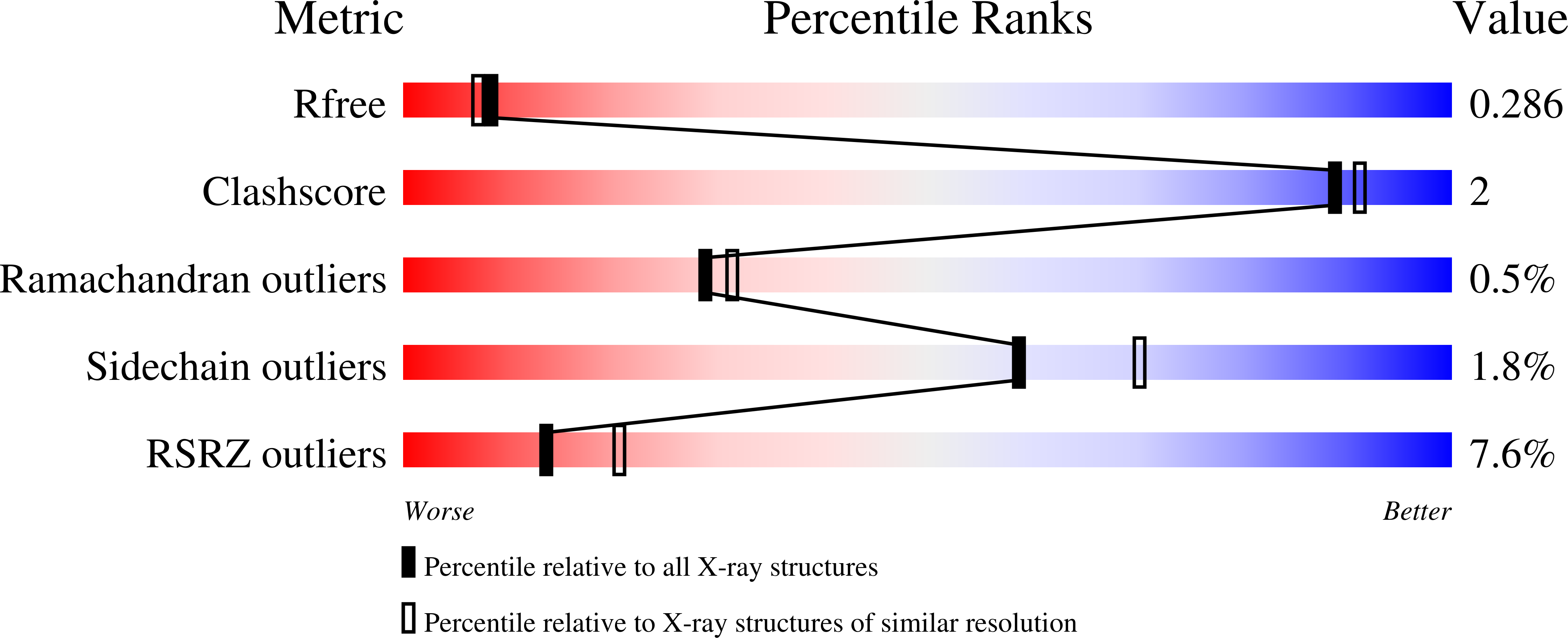
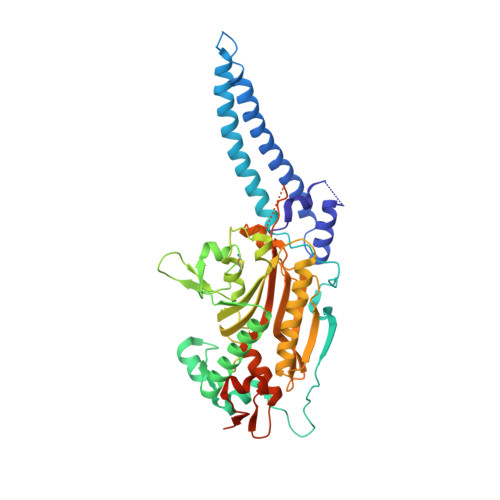
6GIR - PubMed Abstract:
The rules of the genetic code are established by aminoacyl-tRNA synthetases (aaRSs) enzymes, which covalently link tRNA with the cognate amino acid. Many aaRSs are involved in diverse cellular processes beyond translation, acting alone, or in complex with other proteins. However, studies of aaRS noncanonical assembly and functions in plants are scarce, as are structural studies of plant aaRSs. Here, we have solved the crystal structure of Arabidopsis thaliana cytosolic seryl-tRNA synthetase (SerRS), which is the first crystallographic structure of a plant aaRS. Arabidopsis SerRS displays structural features typical of canonical SerRSs, except for a unique intrasubunit disulfide bridge. In a yeast two-hybrid screen, we identified BEN1, a protein involved in the metabolism of plant brassinosteroid hormones, as a protein interactor of Arabidopsis SerRS. The SerRS:BEN1 complex is one of the first protein complexes of plant aaRSs discovered so far, and is a rare example of an aaRS interacting with an enzyme involved in primary or secondary metabolism. To pinpoint regions responsible for this interaction, we created truncated variants of SerRS and BEN1, and identified that the interaction interface involves the SerRS globular catalytic domain and the N-terminal extension of BEN1 protein. BEN1 does not have a strong impact on SerRS aminoacylation activity, indicating that the primary function of the complex is not the modification of SerRS canonical activity. Perhaps SerRS performs as yet unknown noncanonical functions mediated by BEN1. These findings indicate that - via SerRS and BEN1 - a link exists between the protein translation and steroid metabolic pathways of the plant cell. DATABASE: Structural data are available in the PDB under the accession number PDB ID 6GIR.
Organizational Affiliation:
Division of Biochemistry, Department of Chemistry, Faculty of Science, University of Zagreb, Croatia.