Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition.
Robert-Paganin, J., Auguin, D., Houdusse, A.(2018) Nat Commun 9: 4019-4019
- PubMed: 30275503
- DOI: https://doi.org/10.1038/s41467-018-06191-4
- Primary Citation of Related Structures:
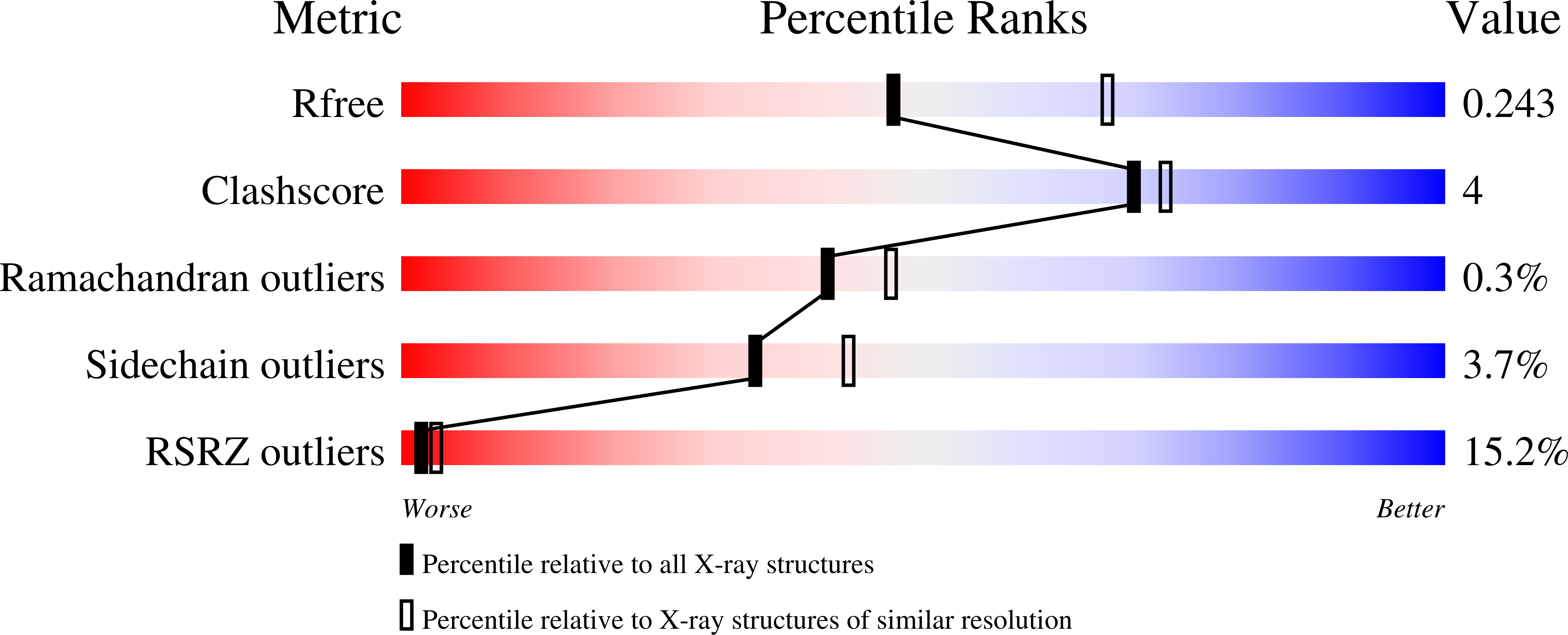
6FSA - PubMed Abstract:
Hypertrophic cardiomyopathies (HCM) result from distinct single-point mutations in sarcomeric proteins that lead to muscle hypercontractility. While different models account for a pathological increase in the power output, clear understanding of the molecular basis of dysfunction in HCM is the mandatory next step to improve current treatments. Here, we present an optimized quasi-atomic model of the sequestered state of cardiac myosin coupled to X-ray crystallography and in silico analysis of the mechanical compliance of the lever arm, allowing the systematic study of a large set of HCM mutations and the definition of different mutation classes based on their effects on lever arm compliance, sequestered state stability, and motor functions. The present work reconciles previous models and explains how distinct HCM mutations can have disparate effects on the motor mechano-chemical parameters and yet lead to the same disease. The framework presented here can guide future investigations aiming at finding HCM treatments.
Organizational Affiliation:
Structural Motility, Institut Curie, PSL Research University, CNRS, UMR 144, F-75005, Paris, France.