Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Banerjee, A., Munir, A., Abdullahu, L., Damha, M.J., Goldgur, Y., Shuman, S.(2019) Nat Commun 10: 218-218
- PubMed: 30644400
- DOI: https://doi.org/10.1038/s41467-018-08211-9
- Primary Citation of Related Structures:
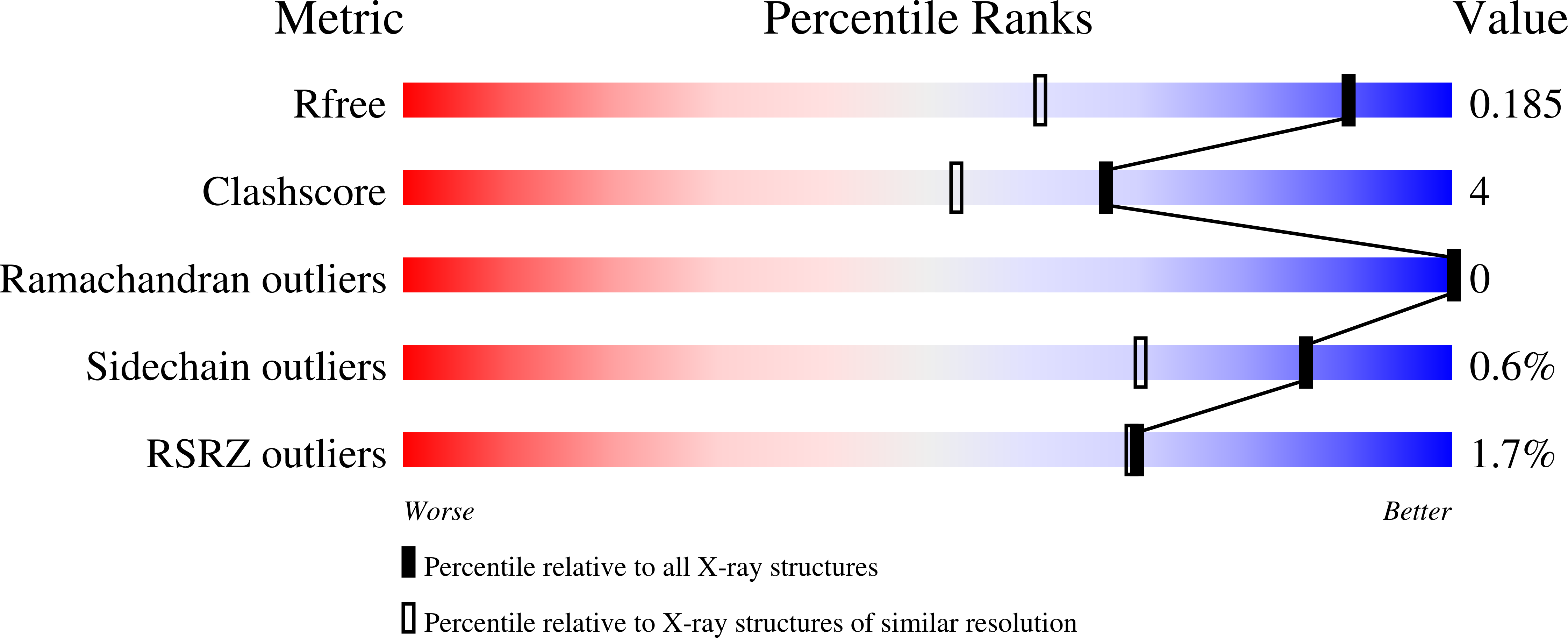
6E3A, 6EDE - PubMed Abstract:
Tpt1 is an essential agent of fungal tRNA splicing that removes the 2'-PO 4 at the splice junction generated by fungal tRNA ligase. Tpt1 catalyzes a unique two-step reaction whereby the 2'-PO 4 attacks NAD + to form an RNA-2'-phospho-ADP-ribosyl intermediate that undergoes transesterification to yield 2'-OH RNA and ADP-ribose-1″,2″-cyclic phosphate products. Because Tpt1 is inessential in exemplary bacterial and mammalian taxa, Tpt1 is seen as an attractive antifungal target. Here we report a 1.4 Å crystal structure of Tpt1 in a product-mimetic complex with ADP-ribose-1″-phosphate in the NAD + site and pAp in the RNA site. The structure reveals how Tpt1 recognizes a 2'-PO 4 RNA splice junction and the mechanism of RNA phospho-ADP-ribosylation. This study also provides evidence that a bacterium has an endogenous phosphorylated substrate with which Tpt1 reacts.
Organizational Affiliation:
Molecular Biology and Structural Biology Programs, Sloan-Kettering Institute, New York, NY, 10065, USA.