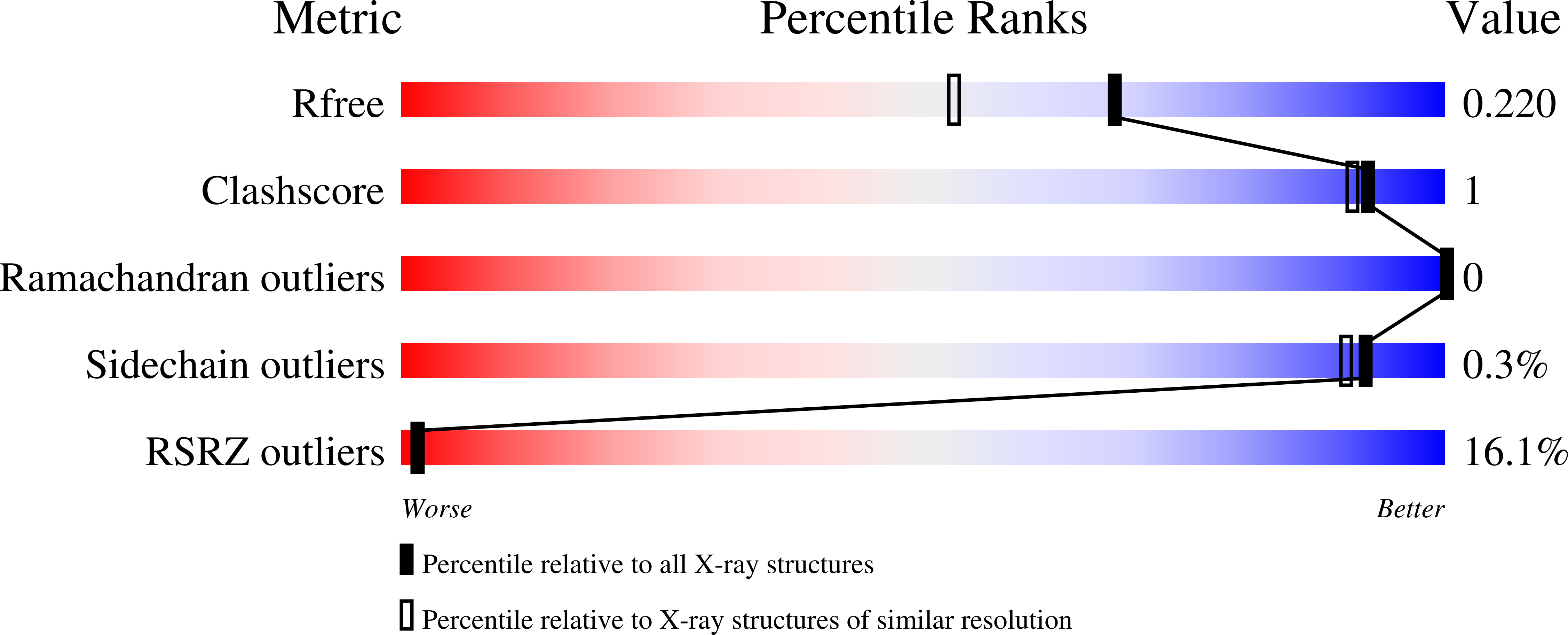
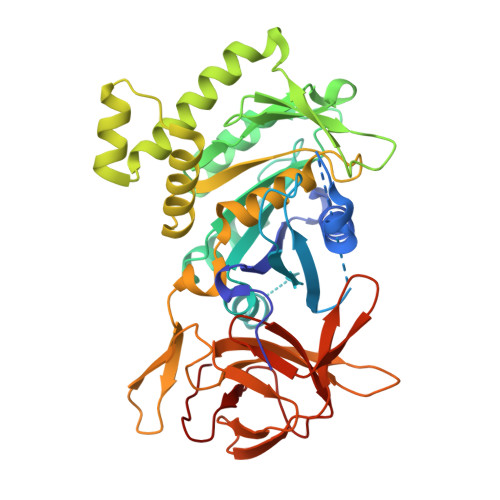
1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
Minasov, G., Shuvalova, L., Dubrovska, I., Kiryukhina, O., Grimshaw, S., Kwon, K., Anderson, W.F., Satchell, K.J.F., Joachimiak, A.To be published.