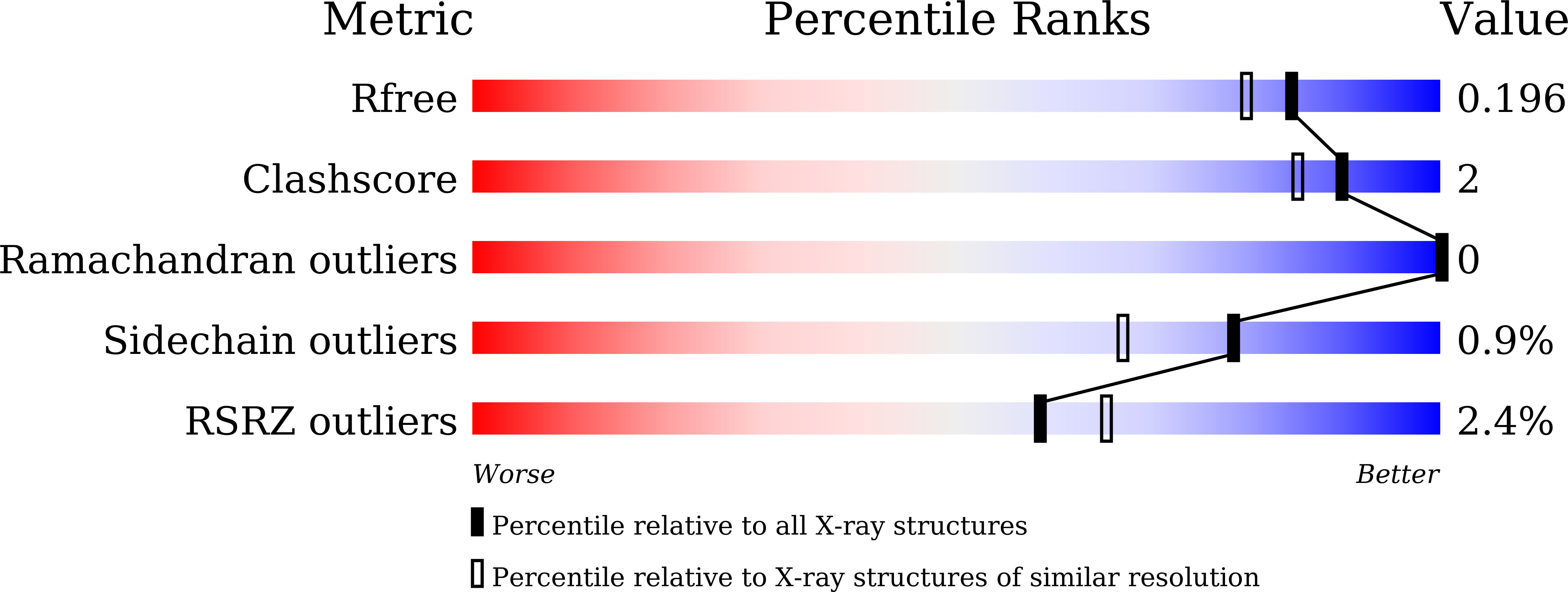
Structural and enzymatic analysis of the cytochrome b5reductase domain of Ulva prolifera nitrate reductase.
You, C., Liu, C., Li, Y., Jiang, P., Ma, Q.(2018) Int J Biol Macromol 111: 1175-1182
- PubMed: 29371148
- DOI: https://doi.org/10.1016/j.ijbiomac.2018.01.140
- Primary Citation of Related Structures:
5YLY - PubMed Abstract:
Rapid accumulations of unattached green macroalgae, referred to as blooms, constitute ecological disasters and occur in many coastal regions. Ulva are a major cause of blooms, owing to their high nitrogen utilization capacity, which requires nitrate reductase (NR) activity; however, molecular characterization of Ulva NR remains lacking. Herein we determined the crystal structure and performed an enzymatic analysis of the cytochrome b 5 reductase domain of Ulva prolifera NR (UpCbRNR). The structural analysis revealed an N-terminal FAD-binding domain primarily consisting of six antiparallel β strands, a C-terminal NADH-binding domain forming a Rossmann fold, and a three β-stranded linker region connecting these two domains. The FAD cofactor was located in the cleft between the two domains and interacted primarily with the FAD-binding domain. UpCbRNR shares similarities in overall structure and cofactor interactions with homologs, and its catalytic ability is comparable to that of higher plant CbRNRs. Structure and sequence comparisons of homologs revealed two regions of sequence length variation potentially useful for phylogenetic analysis: one in the FAD-binding domain, specific to U. prolifera, and another in the linker region that may be used to differentiate between plant, fungi, and animal homologs. Our data will facilitate molecular-level understanding of nitrate assimilation in Ulva.
Organizational Affiliation:
Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Nanhai Road 7, Qingdao 266071, China; Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China; University of Chinese Academy of Sciences, Beijing 100049, China.