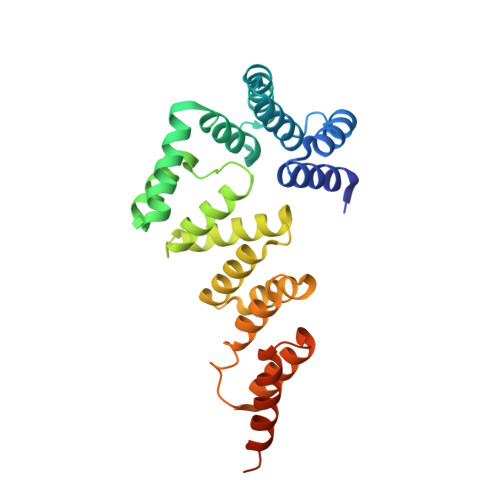
Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C
Nojima, S., Fujishima, A., Kato, K., Ohuchi, K., Shimizu, N., Yonezawa, K., Tajima, K., Yao, M.(2017) Sci Rep 7: 13018-13018
- PubMed: 29026093
- DOI: https://doi.org/10.1038/s41598-017-12530-0
- Primary Citation of Related Structures:
5XW7 - PubMed Abstract:
Bacterial cellulose (BC) is synthesized and exported through the cell membrane via a large protein complex (terminal complex) that consists of three or four subunits. BcsC is a little-studied subunit considered to export BC to the extracellular matrix. It is predicted to have two domains: a tetratrico peptide repeat (TPR) domain and a β-barrelled outer membrane domain. Here we report the crystal structure of the N-terminal part of BcsC-TPR domain (Asp24-Arg272) derived from Enterobacter CJF-002. Unlike most TPR-containing proteins which have continuous TPR motifs, this structure has an extra α-helix between two clusters of TPR motifs. Five independent molecules in the crystal had three different conformations that varied at the hinge of the inserted α-helix. Such structural feature indicates that the inserted α-helix confers flexibility to the chain and changes the direction of the TPR super-helix, which was also suggested by structural analysis of BcsC-TPR (Asp24-Leu664) in solution by size exclusion chromatography-small-angle X-ray scattering. The flexibility at the α-helical hinge may play important role for exporting glucan chains.
Organizational Affiliation:
Graduate school of Life Science, Hokkaido University, Sapporo, Hokkaido, 060-0810, Japan.