Human Argonaute3 has slicer activity.
Park, M.S., Phan, H.D., Busch, F., Hinckley, S.H., Brackbill, J.A., Wysocki, V.H., Nakanishi, K.(2017) Nucleic Acids Res 45: 11867-11877
- PubMed: 29040713
- DOI: https://doi.org/10.1093/nar/gkx916
- Primary Citation of Related Structures:
5VM9 - PubMed Abstract:
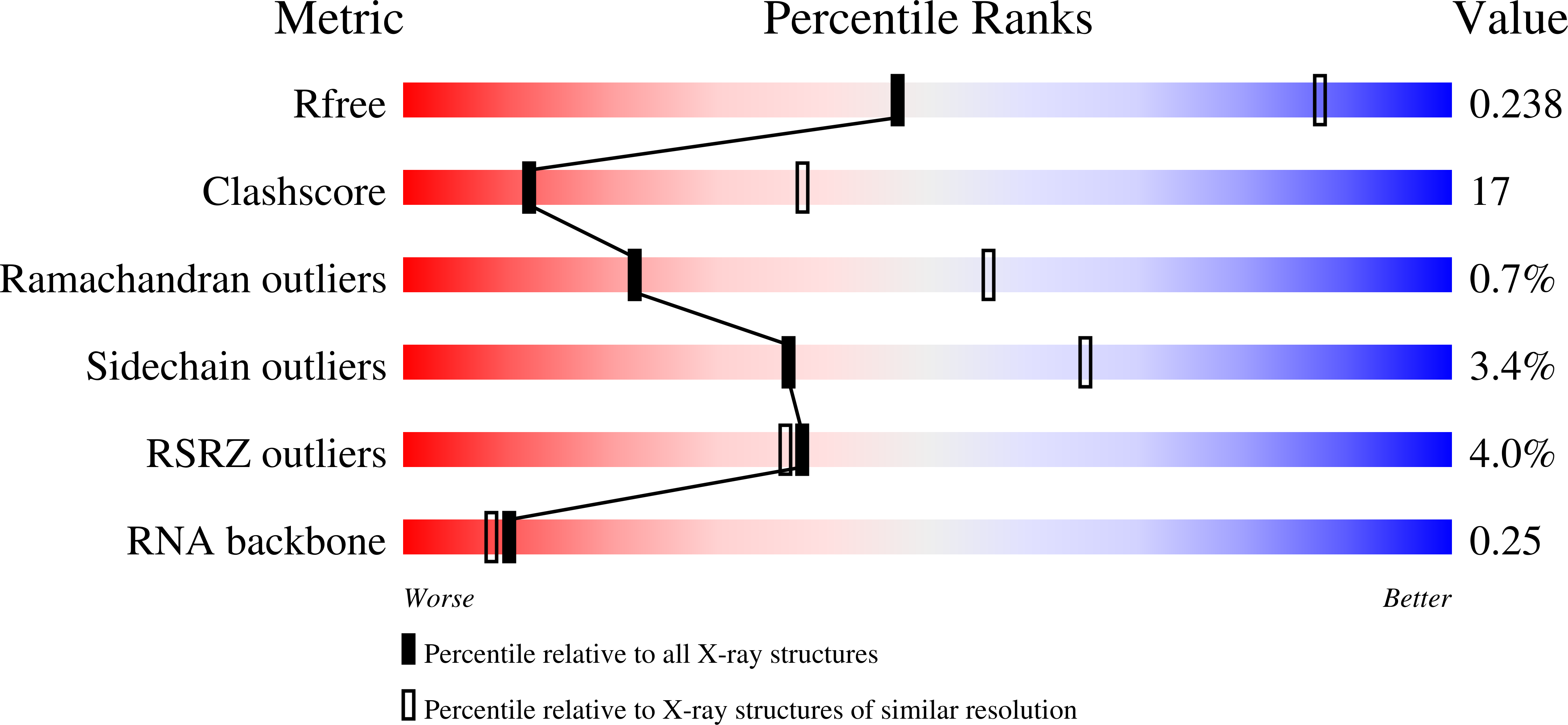
Of the four human Argonaute (AGO) paralogs, only AGO2 has been shown to have slicer activity. The others (AGO1, AGO3 and AGO4) have been thought to assemble with microRNAs to form slicer-independent effector complexes that bind target mRNAs and silence gene expression through translational repression and deadenylation but not cleavage. Here, we report that recombinant AGO3 loaded with miR-20a cleaves complementary target RNAs, whereas AGO3 loaded with let-7a, miR-19b or miR-16 does not, indicating that AGO3 has slicer activity but that this activity depends on the guide RNA. Our cleavage assays using chimeric guides revealed the significance of seed sequence for AGO3 activity, which depends specifically on the sequence of the post-seed. Unlike AGO2, target cleavage by AGO3 requires both 5'- and 3'-flanking regions. Our 3.28 Å crystal structure shows that AGO3 forms a complete active site mirroring that of AGO2, but not a well-defined nucleic acid-binding channel. These results demonstrating that AGO3 also has slicer activity but with more intricate substrate requirements, explain the observation that AGO3 has retained the necessary catalytic residues throughout its evolution. In addition, our structure inspires the idea that the substrate-binding channel of AGO3 and consequently its cellular function, may be modulated by accessory proteins.
Organizational Affiliation:
Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA.