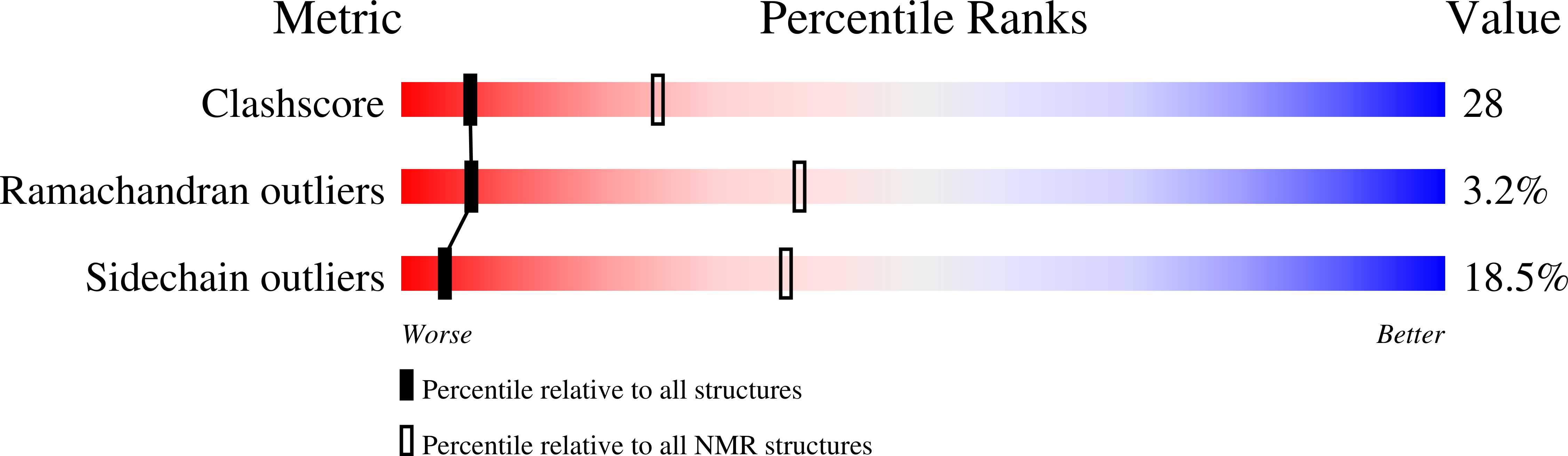NMR structure of the HIV-1 reverse transcriptase thumb subdomain.
Sharaf, N.G., Brereton, A.E., Byeon, I.L., Andrew Karplus, P., Gronenborn, A.M.(2016) J Biomol NMR 66: 273-280
- PubMed: 27858311
- DOI: https://doi.org/10.1007/s10858-016-0077-2
- Primary Citation of Related Structures:
5T82 - PubMed Abstract:
The solution NMR structure of the isolated thumb subdomain of HIV-1 reverse transcriptase (RT) has been determined. A detailed comparison of the current structure with dozens of the highest resolution crystal structures of this domain in the context of the full-length enzyme reveals that the overall structures are very similar, with only two regions exhibiting local conformational differences. The C-terminal capping pattern of the αH helix is subtly different, and the loop connecting the αI and αJ helices in the p51 chain of the full-length p51/p66 heterodimeric RT differs from our NMR structure due to unique packing interactions in mature RT. Overall, our data show that the thumb subdomain folds independently and essentially the same in isolation as in its natural structural context.
Organizational Affiliation:
Department of Structural Biology and Pittsburgh Center for HIV Protein Interactions, University of Pittsburgh, School of Medicine, Biomedical Science Tower 3, 3501 Fifth Avenue, Pittsburgh, PA, 15260, USA.