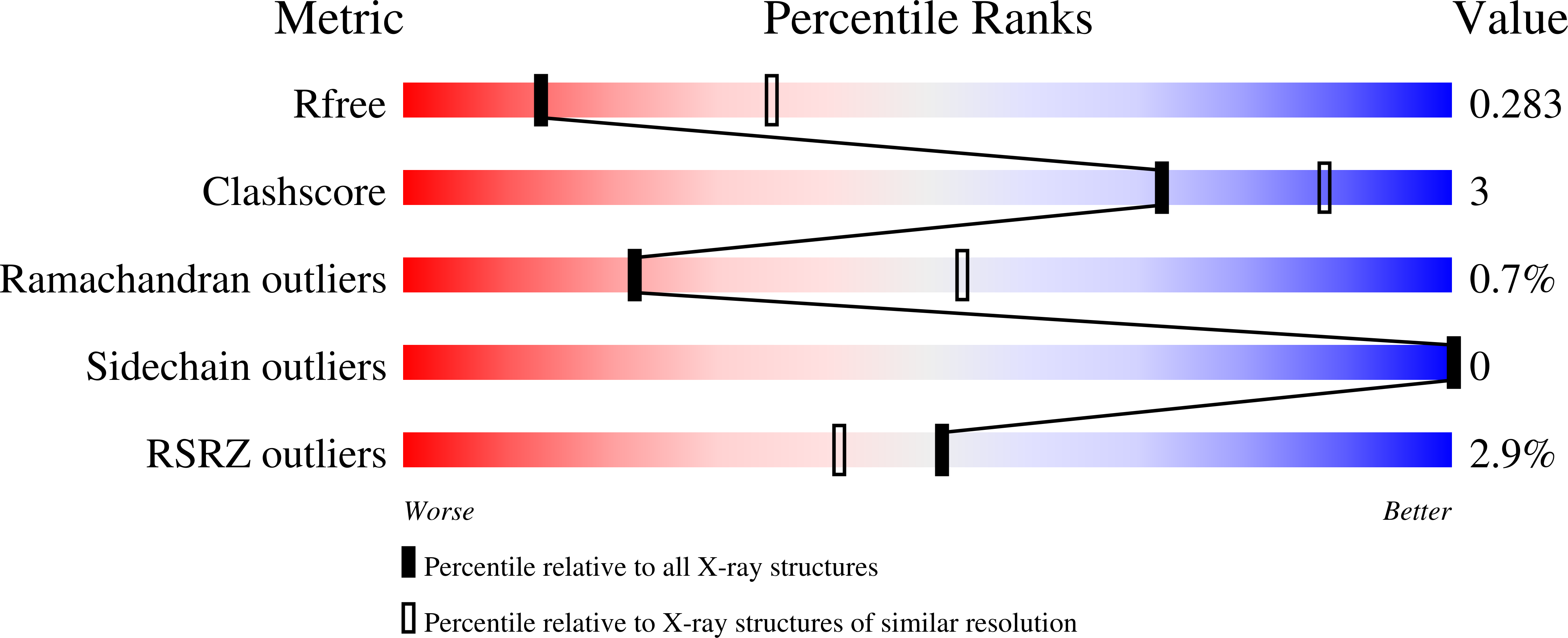
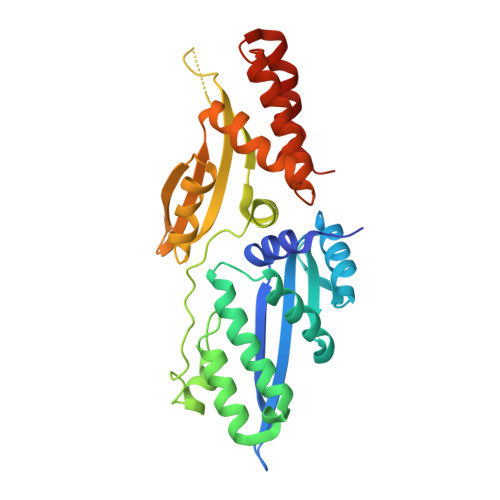
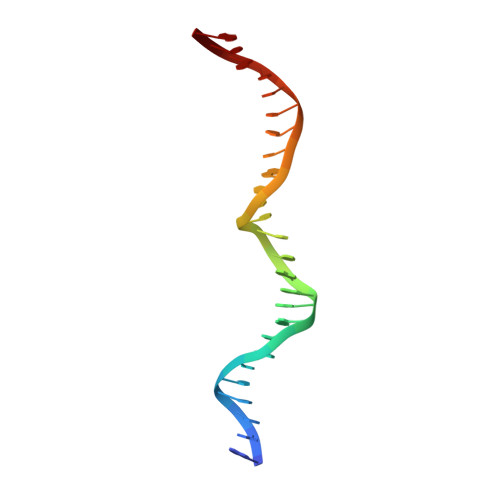
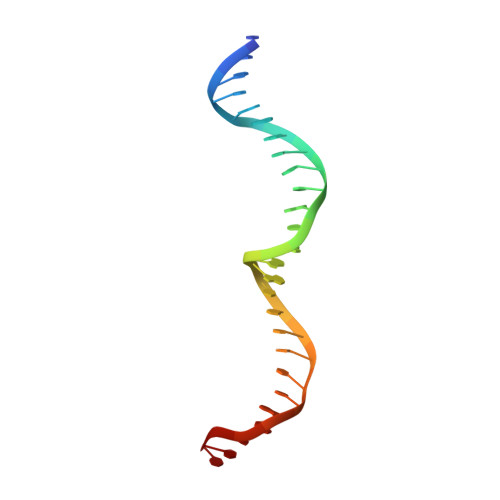
Crystallographic analyses illustrate significant plasticity and efficient recoding of meganuclease target specificity.
Werther, R., Hallinan, J.P., Lambert, A.R., Havens, K., Pogson, M., Jarjour, J., Galizi, R., Windbichler, N., Crisanti, A., Nolan, T., Stoddard, B.L.(2017) Nucleic Acids Res 45: 8621-8634
- PubMed: 28637173
- DOI: https://doi.org/10.1093/nar/gkx544
- Primary Citation of Related Structures:
5T2H, 5T2N, 5T2O - PubMed Abstract:
The retargeting of protein-DNA specificity, outside of extremely modular DNA binding proteins such as TAL effectors, has generally proved to be quite challenging. Here, we describe structural analyses of five different extensively retargeted variants of a single homing endonuclease, that have been shown to function efficiently in ex vivo and in vivo applications. The redesigned proteins harbor mutations at up to 53 residues (18%) of their amino acid sequence, primarily distributed across the DNA binding surface, making them among the most significantly reengineered ligand-binding proteins to date. Specificity is derived from the combined contributions of DNA-contacting residues and of neighboring residues that influence local structural organization. Changes in specificity are facilitated by the ability of all those residues to readily exchange both form and function. The fidelity of recognition is not precisely correlated with the fraction or total number of residues in the protein-DNA interface that are actually involved in DNA contacts, including directional hydrogen bonds. The plasticity of the DNA-recognition surface of this protein, which allows substantial retargeting of recognition specificity without requiring significant alteration of the surrounding protein architecture, reflects the ability of the corresponding genetic elements to maintain mobility and persistence in the face of genetic drift within potential host target sites.
Organizational Affiliation:
Basic Sciences Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave. N., Seattle, WA 98109, USA.