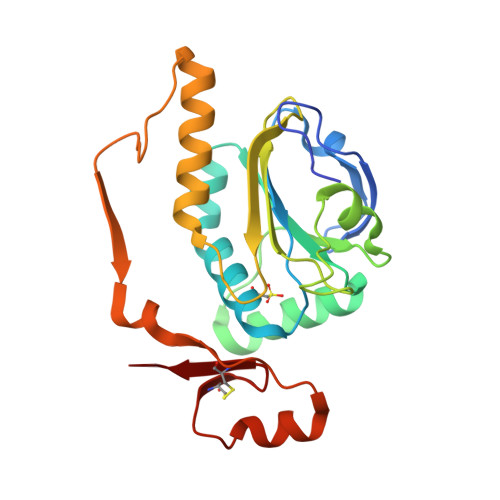
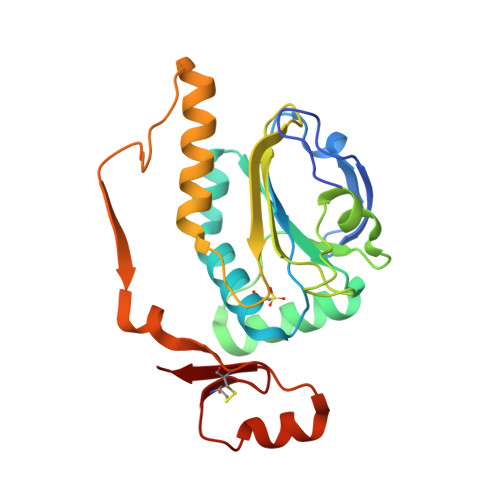
Structural properties of the peroxiredoxin AhpC2 from the hyperthermophilic eubacterium Aquifex aeolicus.
Liu, W., Liu, A., Gao, H., Wang, Q., Wang, L., Warkentin, E., Rao, Z., Michel, H., Peng, G.(2018) Biochim Biophys Acta Gen Subj 1862: 2797-2805
- PubMed: 30251668
- DOI: https://doi.org/10.1016/j.bbagen.2018.08.017
- Primary Citation of Related Structures:
5OVQ - PubMed Abstract:
Peroxiredoxins (Prxs) are thiol peroxidases that scavenge various peroxide substrates such as hydrogen peroxide (H 2 O 2 ), alkyl hydroperoxides and peroxinitrite. They also function as chaperones and are involved in signal transduction by H 2 O 2 in eukaryotic cells. The genome of Aquifex aeolicus, a microaerophilic, hyperthermophilic eubacterium, encodes four Prxs, among them an alkyl hydroperoxide reductase AhpC2 which was found to be closely related to archaeal 1-Cys peroxiredoxins. We determined the crystal structure of AhpC2 at 1.8 Å resolution and investigated its oligomeric state in solution by electron microscopy. AhpC2 is arranged as a toroid-shaped dodecamer instead of the typically observed decamer. The basic folding topology and the active site structure are conserved and possess a high structural similarity to other 1-Cys Prxs. However, the C-terminal region adopts an opposite orientation. AhpC2 contains three cysteines, Cys 49 , Cys 212 , and Cys 218 . The peroxidatic cysteine C P 49 was found to be hyperoxidized to the sulfonic acid (SO 3 H) form, while Cys 212 forms an intra-monomer disulfide bond with Cys 218 . Mutagenesis experiments indicate that Cys 212 and Cys 218 play important roles in the oligomerization of AhpC2. Based on these structural characteristics, we proposed the catalytic mechanism of AhpC2. This study provides novel insights into the structure and reaction mechanism of 1-Cys peroxiredoxins.
Organizational Affiliation:
Department of Molecular Membrane Biology, Max Planck Institute of Biophysics, 60438 Frankfurt am Main, Germany.