Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies.
Garcia-Castellanos, R., Nielsen, N.S., Runager, K., Thgersen, I.B., Lukassen, M.V., Poulsen, E.T., Goulas, T., Enghild, J.J., Gomis-Ruth, F.X.(2017) Structure 25: 1740-1750.e2
- PubMed: 28988748
- DOI: https://doi.org/10.1016/j.str.2017.09.001
- Primary Citation of Related Structures:
5NV6 - PubMed Abstract:
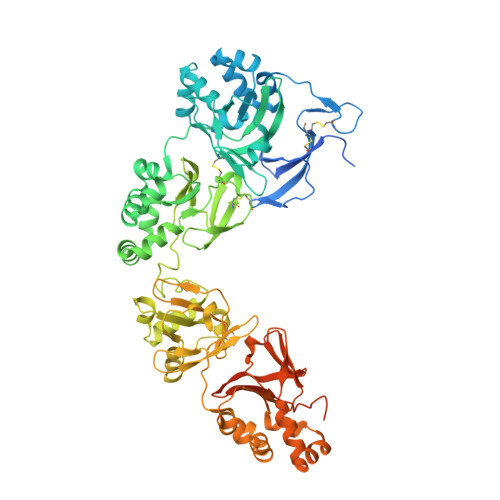
A major cause of visual impairment, corneal dystrophies result from accumulation of protein deposits in the cornea. One of the proteins involved is transforming growth factor β-induced protein (TGFBIp), an extracellular matrix component that interacts with integrins but also produces corneal deposits when mutated. Human TGFBIp is a multi-domain 683-residue protein, which contains one CROPT domain and four FAS1 domains. Its structure spans ∼120 Å and reveals that vicinal domains FAS1-1/FAS1-2 and FAS1-3/FAS1-4 tightly interact in an equivalent manner. The FAS1 domains are sandwiches of two orthogonal four-stranded β sheets decorated with two three-helix insertions. The N-terminal FAS1 dimer forms a compact moiety with the structurally novel CROPT domain, which is a five-stranded all-β cysteine-knot solely found in TGFBIp and periostin. The overall TGFBIp architecture discloses regions for integrin binding and that most dystrophic mutations cluster at both molecule ends, within domains FAS1-1 and FAS1-4.
Organizational Affiliation:
Proteolysis Laboratory, Structural Biology Unit ("María-de-Maeztu" Unit of Excellence), Molecular Biology Institute of Barcelona (CSIC), Barcelona Science Park, c/Baldiri Reixac 15-21, 08028 Barcelona, Catalonia, Spain.