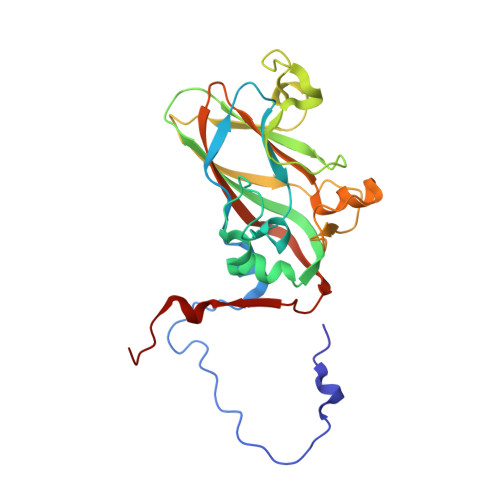
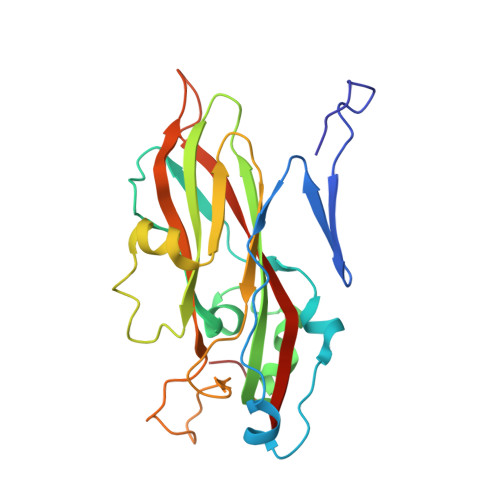
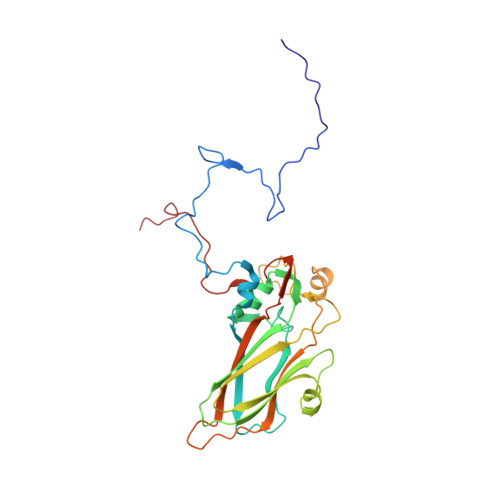
Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Prochazkova, M., Fuzik, T., Skubnik, K., Moravcova, J., Ubiparip, Z., Pridal, A., Plevka, P.(2018) Proc Natl Acad Sci U S A 115: 7759-7764
- PubMed: 29987012
- DOI: https://doi.org/10.1073/pnas.1722018115
- Primary Citation of Related Structures:
5LSF, 5OYP, 6EGV, 6EGX, 6EH1, 6EIW - PubMed Abstract:
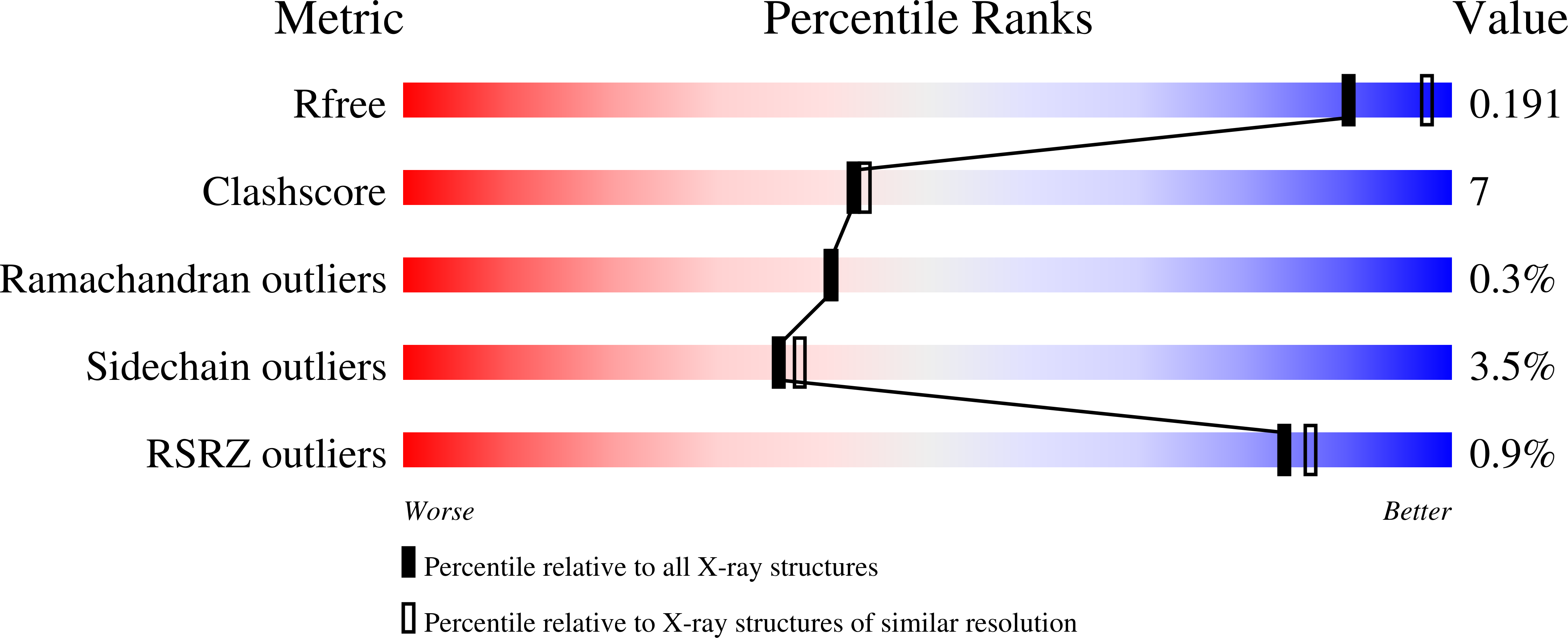
Infection by sacbrood virus (SBV) from the family Iflaviridae is lethal to honey bee larvae but only rarely causes the collapse of honey bee colonies. Despite the negative effect of SBV on honey bees, the structure of its particles and mechanism of its genome delivery are unknown. Here we present the crystal structure of SBV virion and show that it contains 60 copies of a minor capsid protein (MiCP) attached to the virion surface. No similar MiCPs have been previously reported in any of the related viruses from the order Picornavirales. The location of the MiCP coding sequence within the SBV genome indicates that the MiCP evolved from a C-terminal extension of a major capsid protein by the introduction of a cleavage site for a virus protease. The exposure of SBV to acidic pH, which the virus likely encounters during cell entry, induces the formation of pores at threefold and fivefold axes of the capsid that are 7 Å and 12 Å in diameter, respectively. This is in contrast to vertebrate picornaviruses, in which the pores along twofold icosahedral symmetry axes are currently considered the most likely sites for genome release. SBV virions lack VP4 subunits that facilitate the genome delivery of many related dicistroviruses and picornaviruses. MiCP subunits induce liposome disruption in vitro, indicating that they are functional analogs of VP4 subunits and enable the virus genome to escape across the endosome membrane into the cell cytoplasm.
Organizational Affiliation:
Central European Institute of Technology, Masaryk University, 625 00 Brno, Czech Republic.