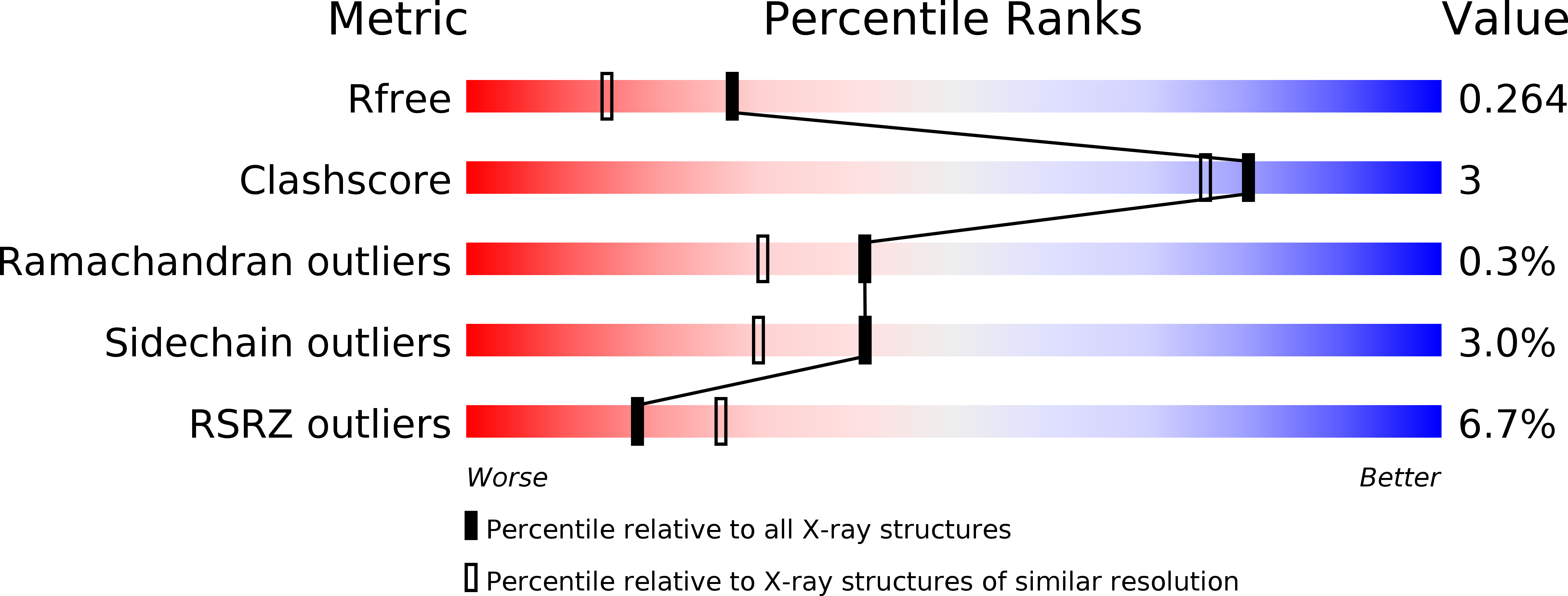
Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
Jiang, Y.L., Jin, H., Yang, H.B., Zhao, R.L., Wang, S., Chen, Y., Zhou, C.Z.(2017) J Biol Chem 292: 6213-6224
- PubMed: 28246170
- DOI: https://doi.org/10.1074/jbc.M116.770446
- Primary Citation of Related Structures:
5GVV, 5GVW - PubMed Abstract:
Protein O -glycosylation is an important post-translational modification in all organisms, but deciphering the specific functions of these glycans is difficult due to their structural complexity. Understanding the glycosylation of mucin-like proteins presents a particular challenge as they are modified numerous times with both the enzymes involved and the glycosylation patterns being poorly understood. Here we systematically explored the O -glycosylation pathway of a mucin-like serine-rich repeat protein PsrP from the human pathogen Streptococcus pneumoniae TIGR4. Previous works have assigned the function of 3 of the 10 glycosyltransferases thought to modify PsrP, GtfA/B, and Gtf3 as catalyzing the first two reactions to form a unified disaccharide core structure. We now use in vivo and in vitro glycosylation assays combined with hydrolytic activity assays to identify the glycosyltransferases capable of decorating this core structure in the third and fourth steps of glycosylation. Specifically, the full-length GlyE and GlyG proteins and the GlyD DUF1792 domain participate in both steps, whereas full-length GlyA and the GlyD GT8 domain catalyze only the fourth step. Incorporation of different sugars to the disaccharide core structure at multiple sites along the serine-rich repeats results in a highly polymorphic product. Furthermore, crystal structures of apo- and UDP-complexed GlyE combined with structural analyses reveal a novel Rossmann-fold "add-on" domain that we speculate to function as a universal module shared by GlyD, GlyE, and GlyA to forward the peptide acceptor from one enzyme to another. These findings define the complete glycosylation pathway of a bacterial glycoprotein and offer a testable hypothesis of how glycosyltransferase coordination facilitates glycan assembly.
Organizational Affiliation:
From the Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China and.