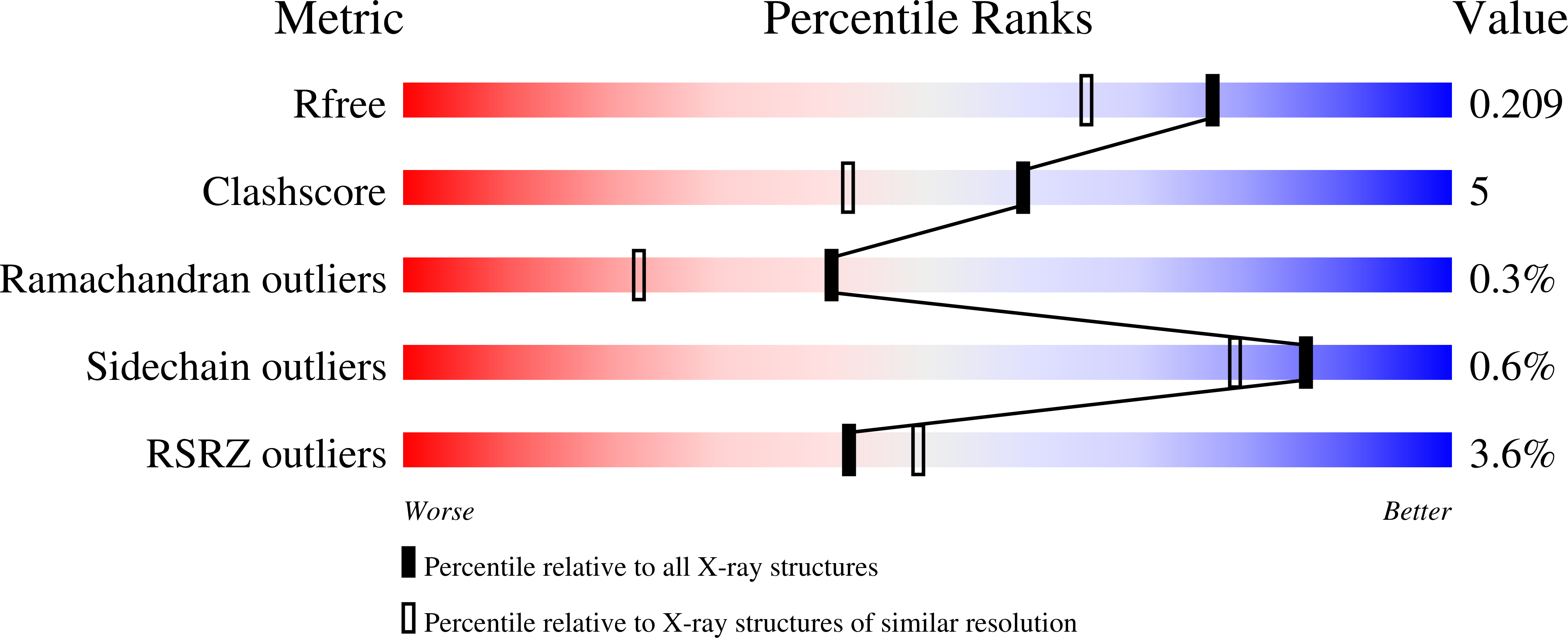
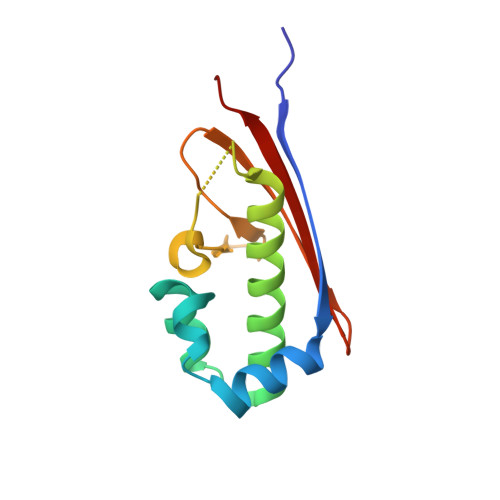
1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
Halavaty, A.S., Minasov, G., Papazisi, L., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.