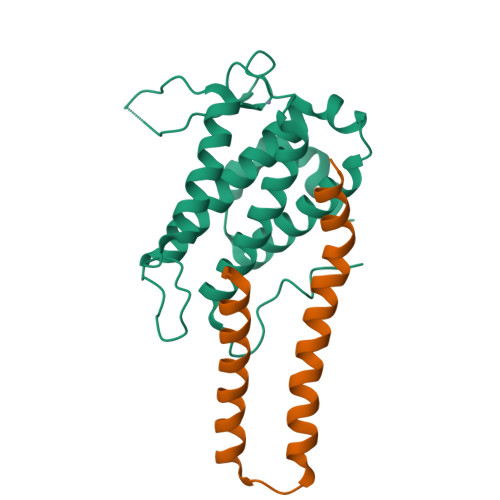
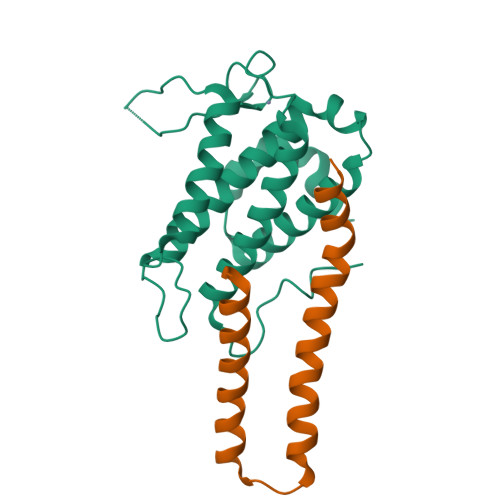
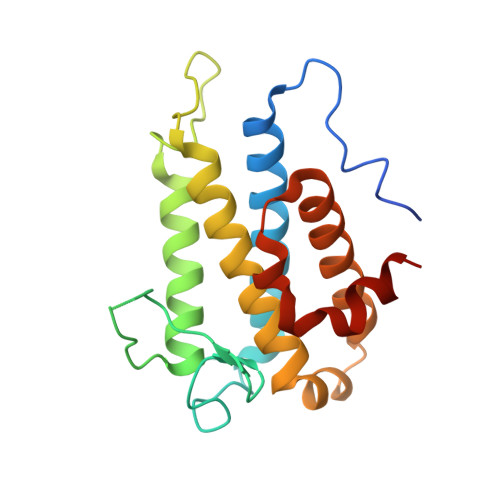
Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Kim, S.Y., Tachioka, Y., Mori, T., Hakoshima, T.(2016) Sci Rep 6: 28488-28488
- PubMed: 27335147
- DOI: https://doi.org/10.1038/srep28488
- Primary Citation of Related Structures:
5B5V, 5B5W, 5B6B - PubMed Abstract:
MOB1 protein is a key regulator of large tumor suppressor 1/2 (LATS1/2) kinases in the Hippo pathway. MOB1 is present in an autoinhibited form and is activated by MST1/2-mediated phosphorylation, although the precise mechanisms responsible for autoinhibition and activation are unknown due to lack of an autoinhibited MOB1 structure. Here, we report on the crystal structure of full-length MOB1B in the autoinhibited form and a complex between the MOB1B core domain and the N-terminal regulation (NTR) domain of LATS1. The structure of full-length MOB1B shows that the N-terminal extension forms a short β-strand, the SN strand, followed by a long conformationally flexible positively-charged linker and α-helix, the Switch helix, which blocks the LATS1 binding surface of MOB1B. The Switch helix is stabilized by β-sheet formation of the SN strand with the S2 strand of the MOB1 core domain. Phosphorylation of Thr12 and Thr35 residues structurally accelerates dissociation of the Switch helix from the LATS1-binding surface by the "pull-the-string" mechanism, thereby enabling LATS1 binding.
Organizational Affiliation:
Structural Biology Laboratory, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.