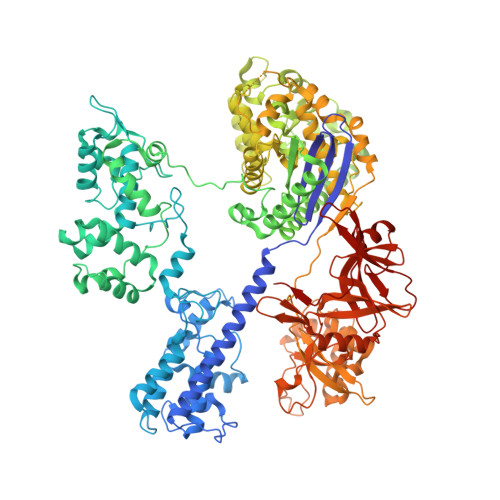
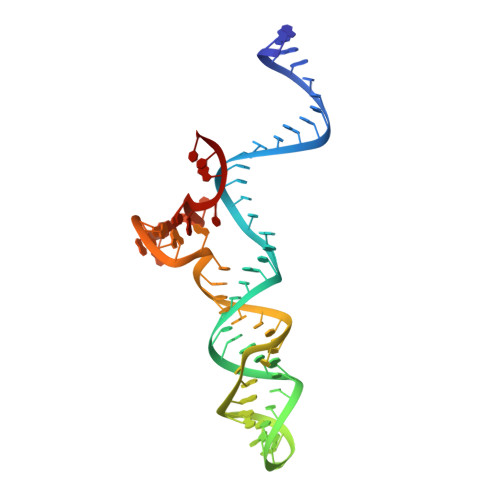
Crystal Structure of Staphylococcus aureus Cas9.
Nishimasu, H., Cong, L., Yan, W.X., Ran, F.A., Zetsche, B., Li, Y., Kurabayashi, A., Ishitani, R., Zhang, F., Nureki, O.(2015) Cell 162: 1113-1126
- PubMed: 26317473
- DOI: https://doi.org/10.1016/j.cell.2015.08.007
- Primary Citation of Related Structures:
5AXW, 5CZZ - PubMed Abstract:
The RNA-guided DNA endonuclease Cas9 cleaves double-stranded DNA targets with a protospacer adjacent motif (PAM) and complementarity to the guide RNA. Recently, we harnessed Staphylococcus aureus Cas9 (SaCas9), which is significantly smaller than Streptococcus pyogenes Cas9 (SpCas9), to facilitate efficient in vivo genome editing. Here, we report the crystal structures of SaCas9 in complex with a single guide RNA (sgRNA) and its double-stranded DNA targets, containing the 5'-TTGAAT-3' PAM and the 5'-TTGGGT-3' PAM, at 2.6 and 2.7 Å resolutions, respectively. The structures revealed the mechanism of the relaxed recognition of the 5'-NNGRRT-3' PAM by SaCas9. A structural comparison of SaCas9 with SpCas9 highlighted both structural conservation and divergence, explaining their distinct PAM specificities and orthologous sgRNA recognition. Finally, we applied the structural information about this minimal Cas9 to rationally design compact transcriptional activators and inducible nucleases, to further expand the CRISPR-Cas9 genome editing toolbox.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; JST, PRESTO, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan.