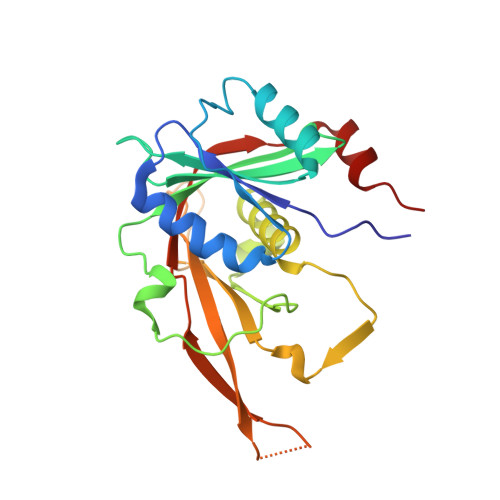
Chaperoning 5S RNA Assembly.
Madru, C., Lebaron, S., Blaud, M., Delbos, L., Pipoli, J., Pasmant, E., Rety, S., Leulliot, N.(2015) Genes Dev 29: 1432
- PubMed: 26159998
- DOI: https://doi.org/10.1101/gad.260349.115
- Primary Citation of Related Structures:
5A53 - PubMed Abstract:
In eukaryotes, three of the four ribosomal RNAs (rRNAs)—the 5.8S, 18S, and 25S/28S rRNAs—are processed from a single pre-rRNA transcript and assembled into ribosomes. The fourth rRNA, the 5S rRNA, is transcribed by RNA polymerase III and is assembled into the 5S ribonucleoprotein particle (RNP), containing ribosomal proteins Rpl5/uL18 and Rpl11/uL5, prior to its incorporation into preribosomes. In mammals, the 5S RNP is also a central regulator of the homeostasis of the tumor suppressor p53. The nucleolar localization of the 5S RNP and its assembly into preribosomes are performed by a specialized complex composed of Rpf2 and Rrs1 in yeast or Bxdc1 and hRrs1 in humans. Here we report the structural and functional characterization of the Rpf2-Rrs1 complex alone, in complex with the 5S RNA, and within pre-60S ribosomes. We show that the Rpf2-Rrs1 complex contains a specialized 5S RNA E-loop-binding module, contacts the Rpl5 protein, and also contacts the ribosome assembly factor Rsa4 and the 25S RNA. We propose that the Rpf2-Rrs1 complex establishes a network of interactions that guide the incorporation of the 5S RNP in preribosomes in the initial conformation prior to its rotation to form the central protuberance found in the mature large ribosomal subunit.
Organizational Affiliation:
Laboratoire de Cristallographie et RMN Biologiques, UMR, CNRS 8015, Université Paris Descartes, Faculté de Pharmacie, Sorbonne Paris Cité, 75006 Paris, France;