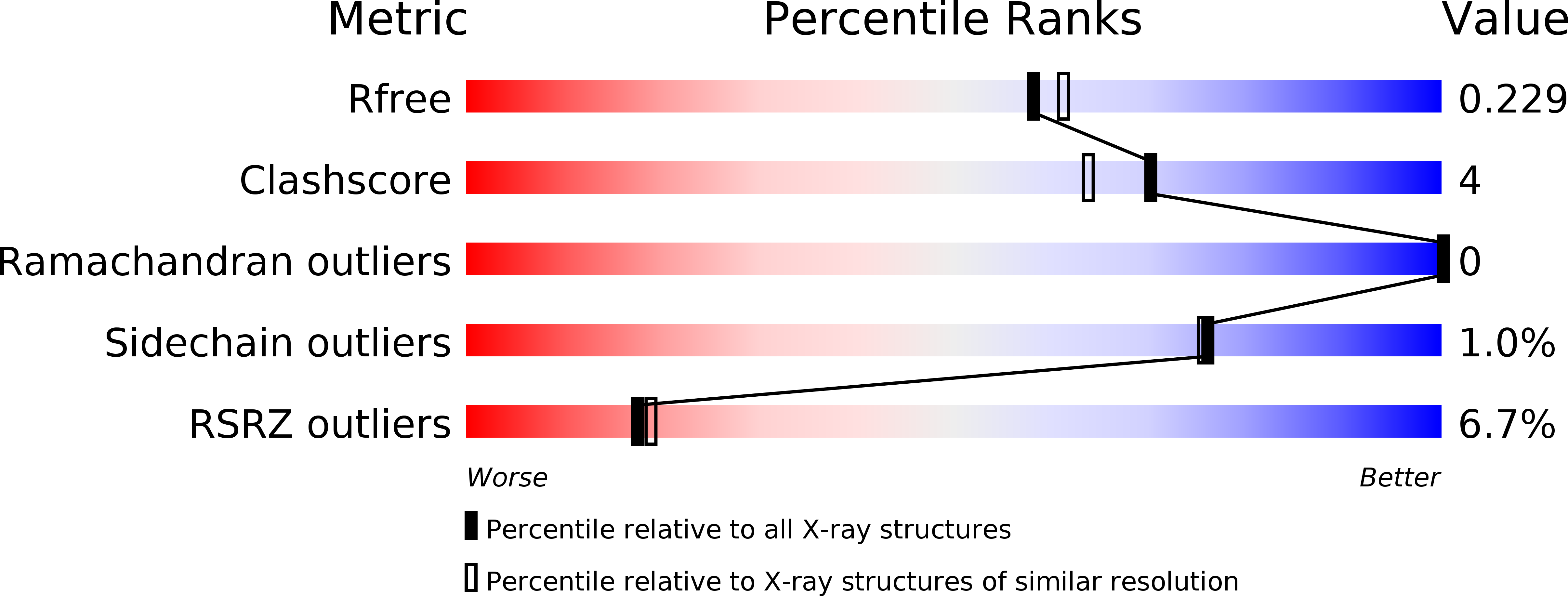
Impact of the crystallization condition on importin-beta conformation.
Tauchert, M.J., Hemonnot, C., Neumann, P., Koster, S., Ficner, R., Dickmanns, A.(2016) Acta Crystallogr D Struct Biol 72: 705-717
- PubMed: 27303791
- DOI: https://doi.org/10.1107/S2059798316004940
- Primary Citation of Related Structures:
4XRI, 4XRK - PubMed Abstract:
In eukaryotic cells, the exchange of macromolecules between the nucleus and cytoplasm is highly selective and requires specialized soluble transport factors. Many of them belong to the importin-β superfamily, the members of which share an overall superhelical structure owing to the tandem arrangement of a specific motif, the HEAT repeat. This structural organization leads to great intrinsic flexibility, which in turn is a prerequisite for the interaction with a variety of proteins and for its transport function. During the passage from the aqueous cytosol into the nucleus, the receptor passes the gated channel of the nuclear pore complex filled with a protein meshwork of unknown organization, which seems to be highly selective owing to the presence of FG-repeats, which are peptides with hydrophobic patches. Here, the structural changes of free importin-β from a single organism, crystallized in polar (salt) or apolar (PEG) buffer conditions, are reported. This allowed analysis of the structural changes, which are attributable to the surrounding milieu and are not affected by bound interaction partners. The importin-β structures obtained exhibit significant conformational changes and suggest an influence of the polarity of the environment, resulting in an extended conformation in the PEG condition. The significance of this observation is supported by SAXS experiments and the analysis of other crystal structures of importin-β deposited in the Protein Data Bank.
Organizational Affiliation:
Department of Molecular Structural Biology, Institute for Microbiology and Genetics, GZMB, Georg-August-University Göttingen, Justus-von-Liebig-Weg 11, 37077 Göttingen, Germany.