Structure and evolution of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27.
Ulrich, A., Wahl, M.C.(2014) Acta Crystallogr D Biol Crystallogr 70: 3110-3123
- PubMed: 25478830
- DOI: https://doi.org/10.1107/S1399004714021695
- Primary Citation of Related Structures:
4R3E, 4R3F - PubMed Abstract:
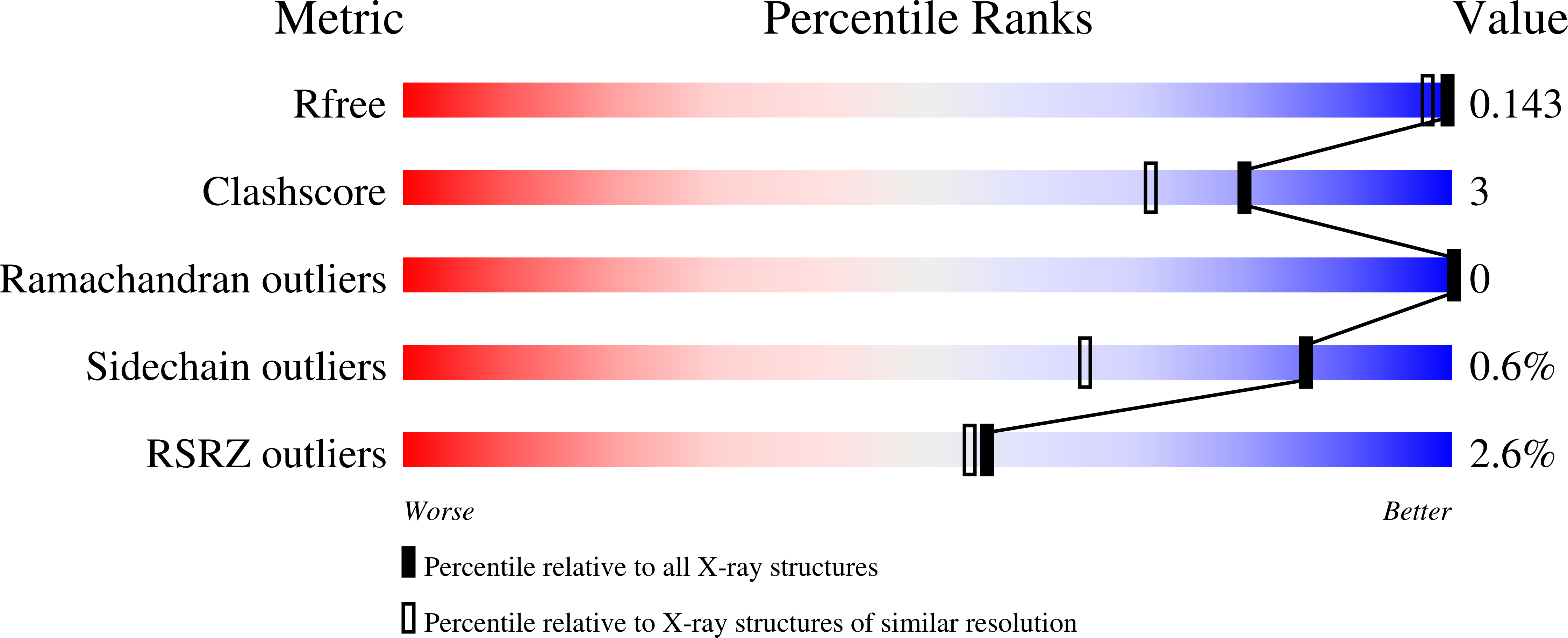
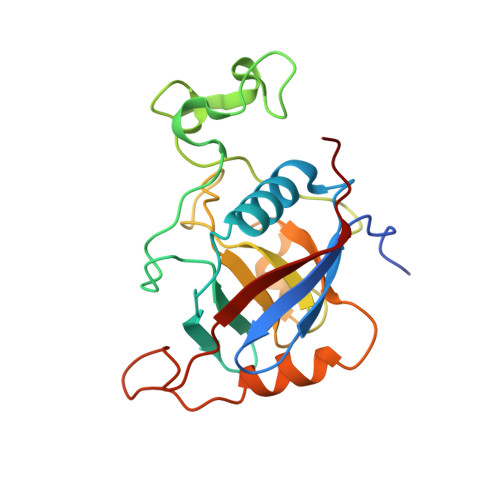
Cwc27 is a spliceosomal cyclophilin-type peptidyl-prolyl cis-trans isomerase (PPIase). Here, the crystal structure of a relatively protease-resistant N-terminal fragment of human Cwc27 containing the PPIase domain was determined at 2.0 Å resolution. The fragment exhibits a C-terminal appendix and resides in a reduced state compared with the previous oxidized structure of a similar fragment. By combining multiple sequence alignments spanning the eukaryotic tree of life and secondary-structure prediction, Cwc27 proteins across the entire eukaryotic kingdom were identified. This analysis revealed the specific loss of a crucial active-site residue in higher eukaryotic Cwc27 proteins, suggesting that the protein evolved from a prolyl isomerase to a pure proline binder. Noting a fungus-specific insertion in the PPIase domain, the 1.3 Å resolution crystal structure of the PPIase domain of Cwc27 from Chaetomium thermophilum was also determined. Although structurally highly similar in the core domain, the C. thermophilum protein displayed a higher thermal stability than its human counterpart, presumably owing to the combined effect of several amino-acid exchanges that reduce the number of long side chains with strained conformations and create new intramolecular interactions, in particular increased hydrogen-bond networks.
Organizational Affiliation:
Laboratory of Structural Biochemistry, Freie Universität Berlin, Takustrasse 6, 14195 Berlin, Germany.