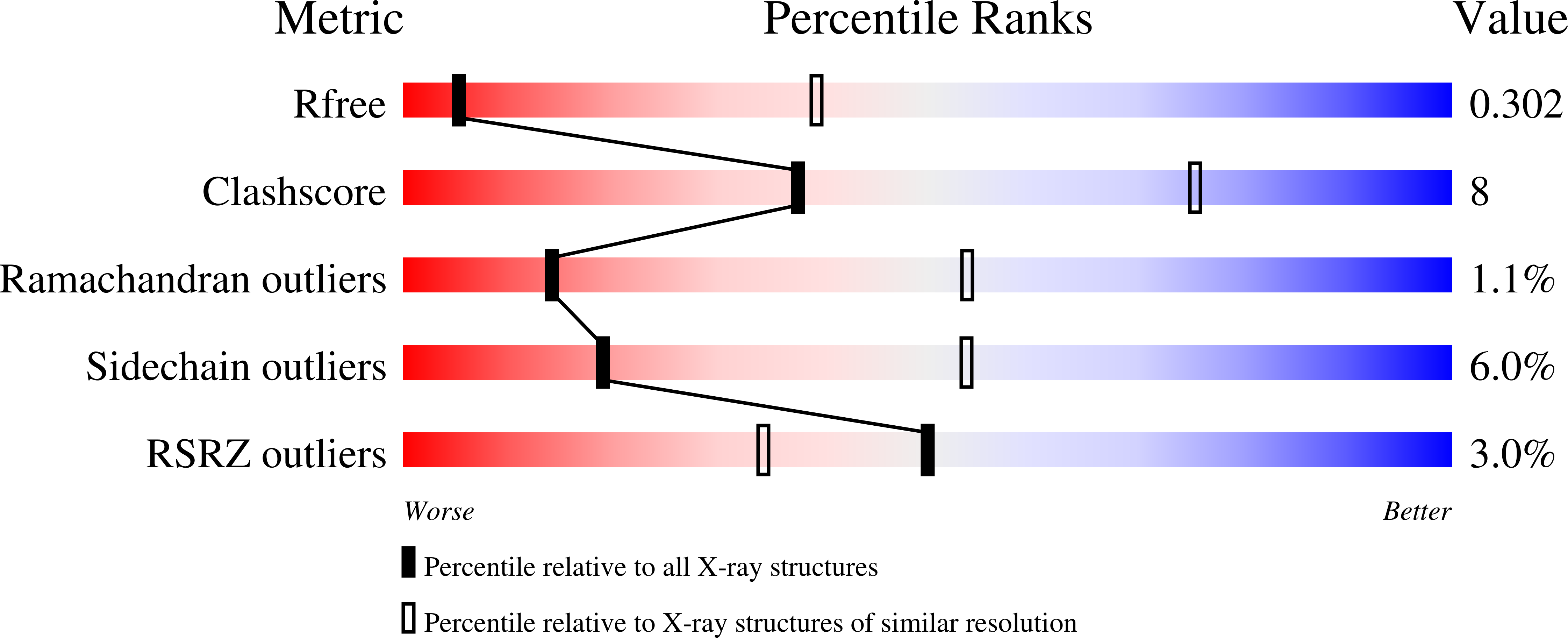
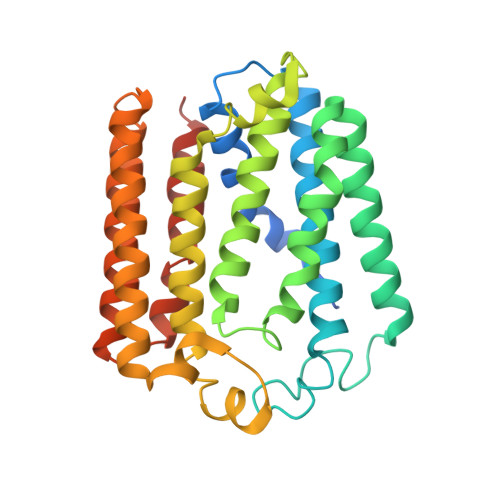
Structural insights into ubiquinone biosynthesis in membranes.
Cheng, W., Li, W.(2014) Science 343: 878-881
- PubMed: 24558159
- DOI: https://doi.org/10.1126/science.1246774
- Primary Citation of Related Structures:
4OD4, 4OD5 - PubMed Abstract:
Biosynthesis of ubiquinones requires the intramembrane UbiA enzyme, an archetypal member of a superfamily of prenyltransferases that generates lipophilic aromatic compounds. Mutations in eukaryotic superfamily members have been linked to cardiovascular degeneration and Parkinson's disease. To understand how quinones are produced within membranes, we report the crystal structures of an archaeal UbiA in its apo and substrate-bound states at 3.3 and 3.6 angstrom resolution, respectively. The structures reveal nine transmembrane helices and an extramembrane cap domain that surround a large central cavity containing the active site. To facilitate the catalysis inside membranes, UbiA has an unusual active site that opens laterally to the lipid bilayer. Our studies illuminate general mechanisms for substrate recognition and catalysis in the UbiA superfamily and rationalize disease-related mutations in humans.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, USA.