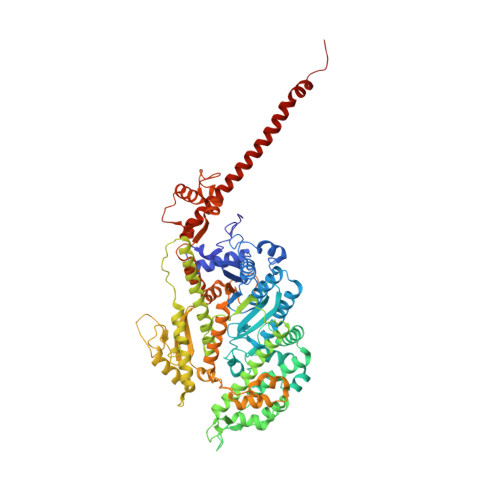
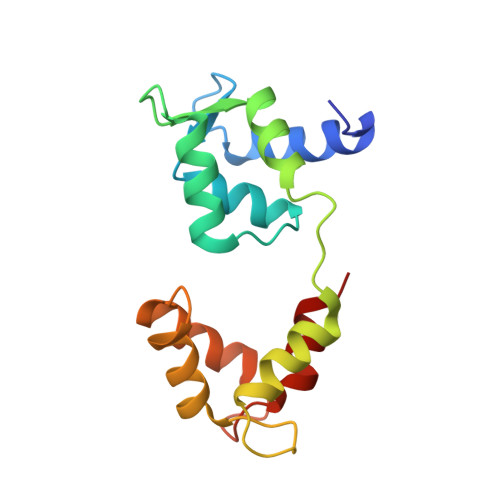
A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Shuman, H., Greenberg, M.J., Zwolak, A., Lin, T., Sindelar, C.V., Dominguez, R., Ostap, E.M.(2014) Proc Natl Acad Sci U S A 111: 2116-2121
- PubMed: 24469830
- DOI: https://doi.org/10.1073/pnas.1321022111
- Primary Citation of Related Structures:
4L79 - PubMed Abstract:
Myosins are molecular motors that power diverse cellular processes, such as rapid organelle transport, muscle contraction, and tension-sensitive anchoring. The structural adaptations in the motor that allow for this functional diversity are not known, due, in part, to the lack of high-resolution structures of highly tension-sensitive myosins. We determined a 2.3-Å resolution structure of apo-myosin-Ib (Myo1b), which is the most tension-sensitive myosin characterized. We identified a striking unique orientation of structural elements that position the motor's lever arm. This orientation results in a cavity between the motor and lever arm that holds a 10-residue stretch of N-terminal amino acids, a region that is divergent among myosins. Single-molecule and biochemical analyses show that the N terminus plays an important role in stabilizing the post power-stroke conformation of Myo1b and in tuning the rate of the force-sensitive transition. We propose that this region plays a general role in tuning the mechanochemical properties of myosins.
Organizational Affiliation:
Pennsylvania Muscle Institute and Department of Physiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104.