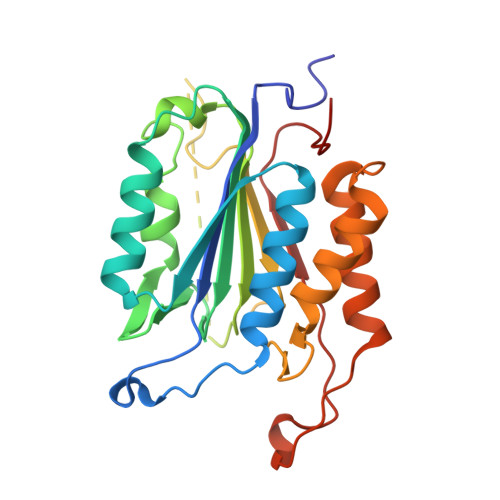
Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Thomsen, N.D., Koerber, J.T., Wells, J.A.(2013) Proc Natl Acad Sci U S A 110: 8477-8482
- PubMed: 23650375
- DOI: https://doi.org/10.1073/pnas.1306759110
- Primary Citation of Related Structures:
4JQY, 4JQZ, 4JR0, 4JR1, 4JR2 - PubMed Abstract:
Procaspase-3 (P3) and procaspase-7 (P7) are activated through proteolytic maturation to form caspase-3 (C3) and caspase-7 (C7), respectively, which serve overlapping but nonredundant roles as the executioners of apoptosis in humans. However, it is unclear if differences in P3 and P7 maturation mechanisms underlie their unique biological functions, as the structure of P3 remains unknown. Here, we report structures of P3 in a catalytically inactive conformation, structures of P3 and P7 bound to covalent peptide inhibitors that reveal the active conformation of the zymogens, and the structure of a partially matured C7:P7 heterodimer. Along with a biochemical analysis, we show that P3 is catalytically inactive and matures through a symmetric all-or-nothing process. In contrast, P7 contains latent catalytic activity and matures through an asymmetric and tiered mechanism, suggesting a lower threshold for activation. Finally, we use our structures to design a selection strategy for conformation specific antibody fragments that stimulate procaspase activity, showing that executioner procaspase conformational equilibrium can be rationally modulated. Our studies provide a structural framework that may help to explain the unique roles of these important proapoptotic enzymes, and suggest general strategies for the discovery of proenzyme activators.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94158, USA.