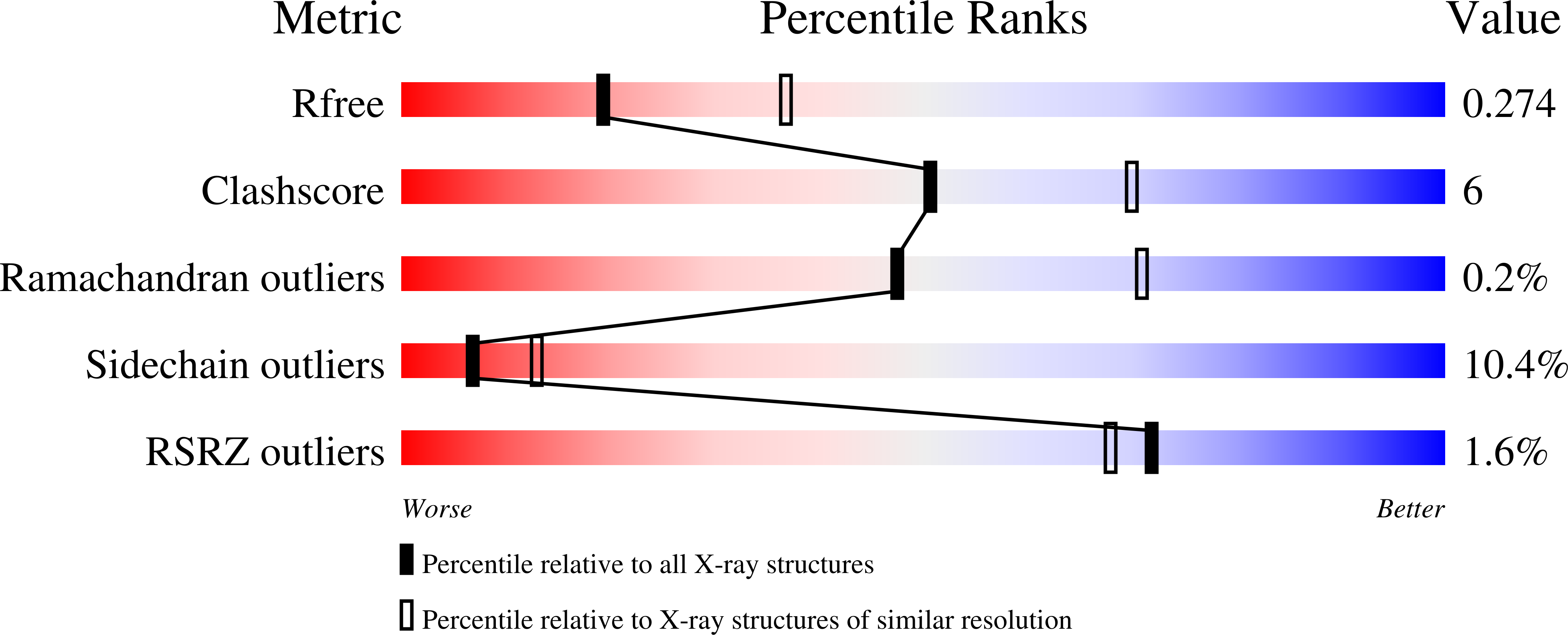
Mechanistic and structural basis for inhibition of thymidylate synthase ThyX.
Basta, T., Boum, Y., Briffotaux, J., Becker, H.F., Lamarre-Jouenne, I., Lambry, J.C., Skouloubris, S., Liebl, U., Graille, M., van Tilbeurgh, H., Myllykallio, H.(2012) Open Biol 2: 120120-120120
- PubMed: 23155486
- DOI: https://doi.org/10.1098/rsob.120120
- Primary Citation of Related Structures:
4FZB - PubMed Abstract:
Nature has established two mechanistically and structurally unrelated families of thymidylate synthases that produce de novo thymidylate or dTMP, an essential DNA precursor. Representatives of the alternative flavin-dependent thymidylate synthase family, ThyX, are found in a large number of microbial genomes, but are absent in humans. We have exploited the nucleotide binding pocket of ThyX proteins to identify non-substrate-based tight-binding ThyX inhibitors that inhibited growth of genetically modified Escherichia coli cells dependent on thyX in a manner mimicking a genetic knockout of thymidylate synthase. We also solved the crystal structure of a viral ThyX bound to 2-hydroxy-3-(4-methoxybenzyl)-1,4-naphthoquinone at a resolution of 2.6 Å. This inhibitor was found to bind within the conserved active site of the tetrameric ThyX enzyme, at the interface of two monomers, partially overlapping with the dUMP binding pocket. Our studies provide new chemical tools for investigating the ThyX reaction mechanism and establish a novel mechanistic and structural basis for inhibition of thymidylate synthesis. As essential ThyX proteins are found e.g. in Mycobacterium tuberculosis and Helicobacter pylori, our studies have also potential to pave the way towards the development of new anti-microbial compounds.
Organizational Affiliation:
Laboratoire d'Optique et Biosciences, INSERM U696, CNRS UMR 7645, Ecole Polytechnique, Palaiseau Cedex, Palaiseau 91228, France.