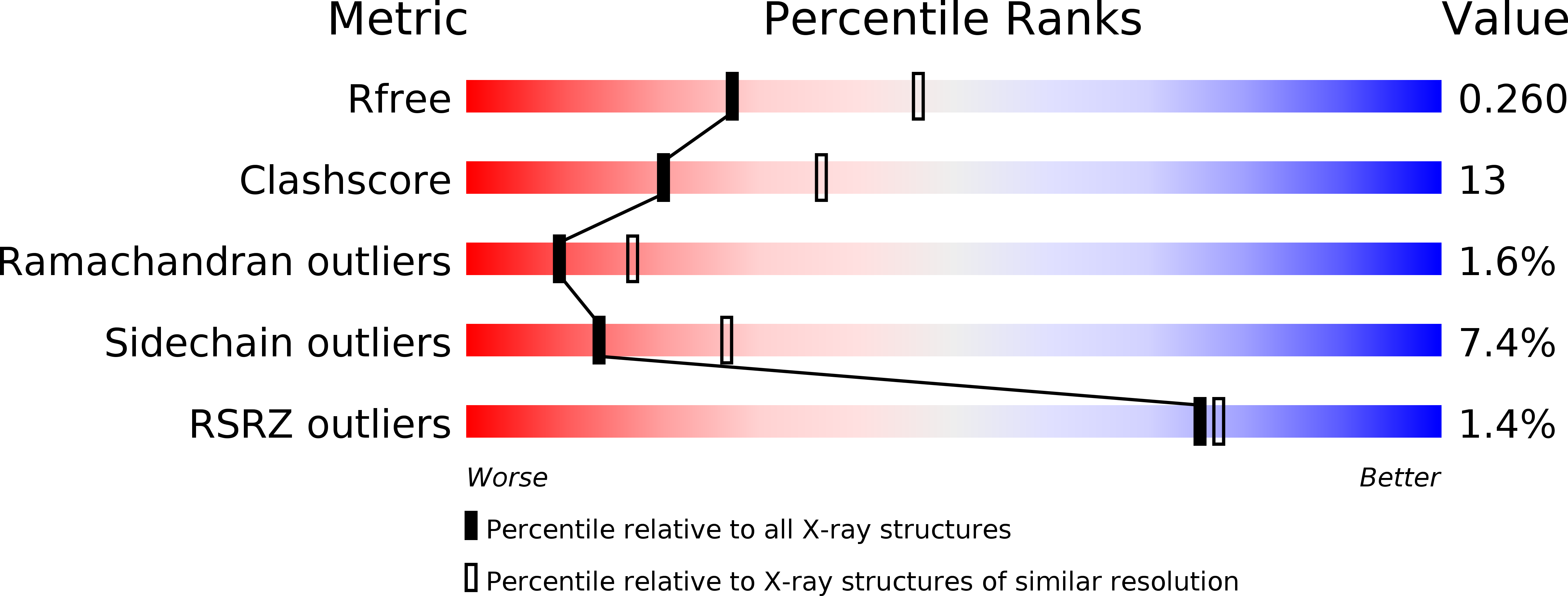
The 2.5 A Structure of the Enterococcus Conjugation Protein TraM resembles VirB8 Type IV Secretion Proteins.
Goessweiner-Mohr, N., Grumet, L., Arends, K., Pavkov-Keller, T., Gruber, C.C., Gruber, K., Birner-Gruenberger, R., Kropec-Huebner, A., Huebner, J., Grohmann, E., Keller, W.(2013) J Biol Chem 288: 2018-2028
- PubMed: 23188825
- DOI: https://doi.org/10.1074/jbc.M112.428847
- Primary Citation of Related Structures:
4EC6 - PubMed Abstract:
Conjugative plasmid transfer is the most important means of spreading antibiotic resistance and virulence genes among bacteria and therefore presents a serious threat to human health. The process requires direct cell-cell contact made possible by a multiprotein complex that spans cellular membranes and serves as a channel for macromolecular secretion. Thus far, well studied conjugative type IV secretion systems (T4SS) are of Gram-negative (G-) origin. Although many medically relevant pathogens (e.g., enterococci, staphylococci, and streptococci) are Gram-positive (G+), their conjugation systems have received little attention. This study provides structural information for the transfer protein TraM of the G+ broad host range Enterococcus conjugative plasmid pIP501. Immunolocalization demonstrated that the protein localizes to the cell wall. We then used opsonophagocytosis as a novel tool to verify that TraM was exposed on the cell surface. In these assays, antibodies generated to TraM recruited macrophages and enabled killing of pIP501 harboring Enteroccocus faecalis cells. The crystal structure of the C-terminal, surface-exposed domain of TraM was determined to 2.5 Å resolution. The structure, molecular dynamics, and cross-linking studies indicated that a TraM trimer acts as the biological unit. Despite the absence of sequence-based similarity, TraM unexpectedly displayed a fold similar to the T4SS VirB8 proteins from Agrobacterium tumefaciens and Brucella suis (G-) and to the transfer protein TcpC from Clostridium perfringens plasmid pCW3 (G+). Based on the alignments of secondary structure elements of VirB8-like proteins from mobile genetic elements and chromosomally encoded T4SS from G+ and G- bacteria, we propose a new classification scheme of VirB8-like proteins.
Organizational Affiliation:
Karl-Franzens-University Graz, Institute of Molecular Biosciences, Structural Biology, 8010 Graz, Austria.