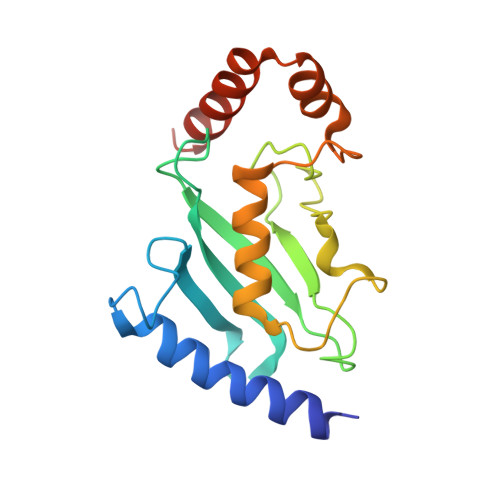
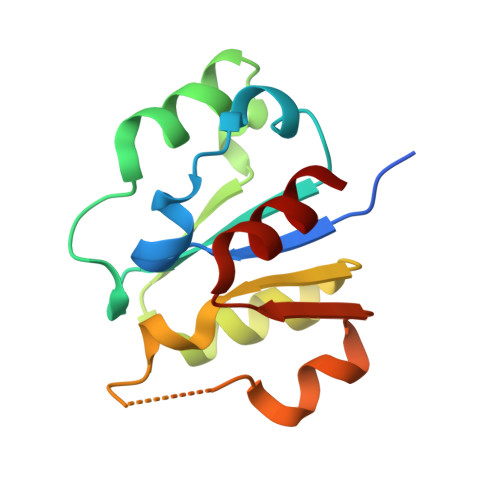
A Disulphide Bond in the E2 Enzyme Pex4P Modulates Ubiquitin-Conjugating Activity
Williams, C., van den Berg, M., Stanley, W.A., Wilmanns, M., Distel, B.(2013) Sci Rep 3: 2212
- PubMed: 23896733
- DOI: https://doi.org/10.1038/srep02212
- Primary Citation of Related Structures:
4BWF - PubMed Abstract:
The ubiquitin-conjugating enzyme Pex4p together with its binding partner, the peroxisomal membrane protein Pex22p, co-ordinates cysteine-dependent ubiquitination of the cycling receptor protein Pex5p. Unusually for an ubiquitin-conjugating enzyme, Saccharomyces cerevisiae Pex4p can form a disulphide bond between the cysteine residues at positions 105 and 146. We found that mutating the disulphide forming cysteine residues in Pex4p to serines does not disturb the secondary structure of the protein but does reduce the in vitro activity of Pex4p. From the crystal structure of Pex4p C105S, C146S in complex with the soluble domain of Pex22p, we observe a narrowing of the active site cleft, caused by loss of the disulphide bond. This modification of the active site microenvironment is likely to restrict access of ubiquitin to the active site cysteine, modulating Pex4p activity. Finally, based on sequence and structural alignments, we have identified other ubiquitin-conjugating enzymes that may contain disulphide bonds.
Organizational Affiliation:
European Molecular Biology Laboratory, Structural Biology Unit, Notkestrasse 85, 22603, Hamburg, Germany.