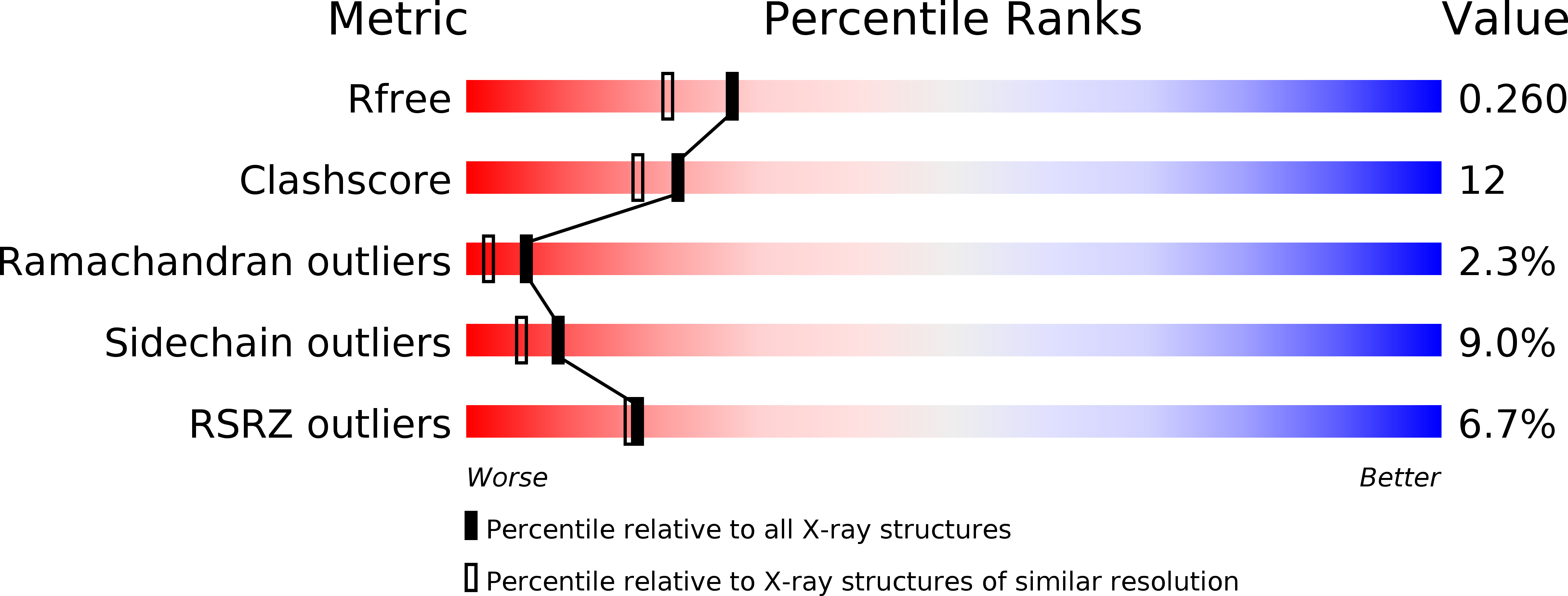
Trim21 is an Igg Receptor that is Structurally, Thermodynamically, and Kinetically Conserved.
Keeble, A.H., Khan, Z., Forster, A., James, L.C.(2008) Proc Natl Acad Sci U S A 105: 6045
- PubMed: 18420815
- DOI: https://doi.org/10.1073/pnas.0800159105
- Primary Citation of Related Structures:
2VOK, 3ZO0 - PubMed Abstract:
The newly identified tripartite motif (TRIM) family of proteins mediate innate immunity and other critical cellular functions. Here we show that TRIM21, which mediates the autoimmune diseases rheumatoid arthritis, systemic lupus erythematosus, and Sjögren's syndrome, is a previously undescribed IgG receptor with a binding mechanism unlike known mammalian Fcgamma receptors. TRIM21 simultaneously targets conserved hot-spot residues on both Ig domains of the Fc fragment using a PRYSPRY domain with a preformed multisite interface. The binding sites on both TRIM21 and Fc are highly conserved to the extent that the proteins are functionally interchangeable through murine, canine, primate, and human species. Pre-steady-state analysis exposes mechanistic conservation at the level of individual residues, which make the same energetic and kinetic contributions to binding despite varying in sequence. Together, our results reveal that TRIM21 is a previously undescribed type of IgG receptor based on a non-Ig scaffold whose interaction at the fundamental level-structural, thermodynamic, and kinetic-is evolutionarily conserved.
Organizational Affiliation:
Division of Protein and Nucleic Acid Chemistry, Laboratory of Molecular Biology, Medical Research Council, Hills Road, Cambridge CB2 2QH, United Kingdom.