Snapshots of a protein folding intermediate
Yamada, S., Bouley-Ford, N.D., Keller, G.E., Ford, W.C., Gray, H.B., Winkler, J.R.(2013) Proc Natl Acad Sci U S A 110: 1606-1610
- PubMed: 23319660
- DOI: https://doi.org/10.1073/pnas.1221832110
- Primary Citation of Related Structures:
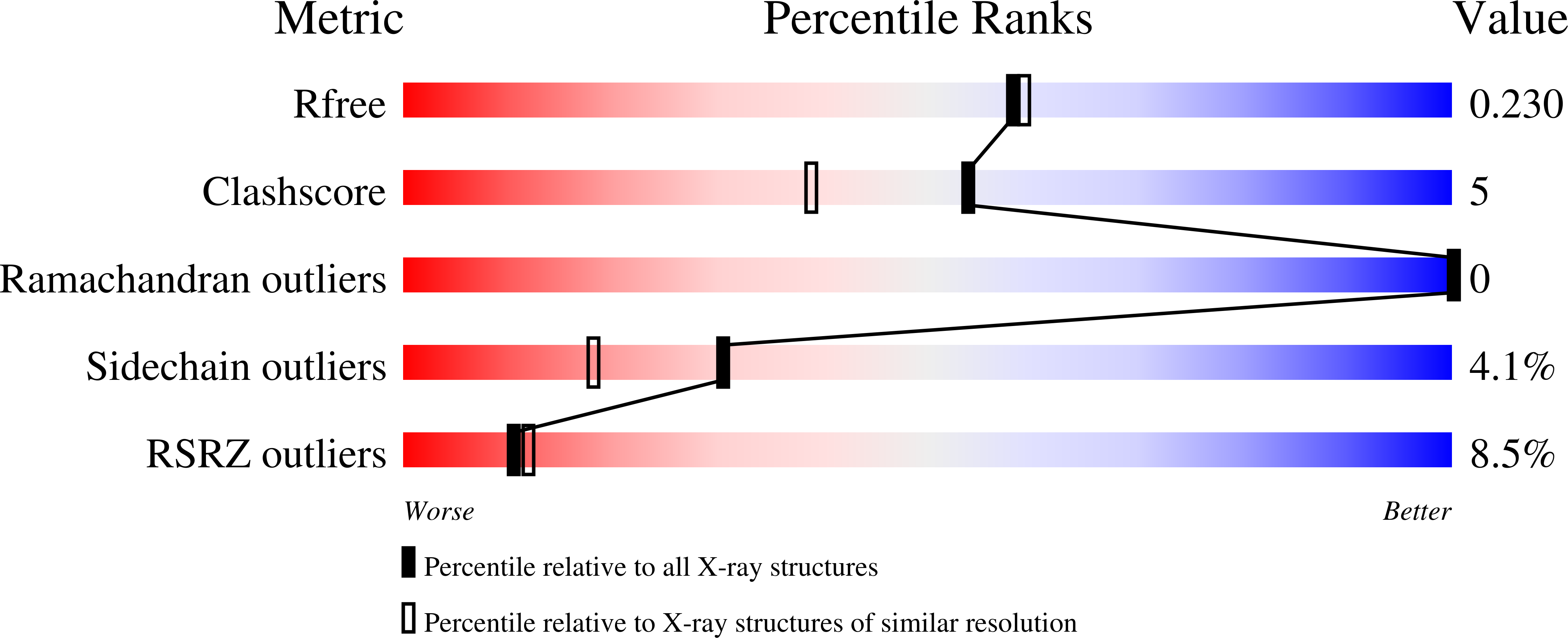
3VNW - PubMed Abstract:
We have investigated the folding dynamics of Thermus thermophilus cytochrome c(552) by time-resolved fluorescence energy transfer between the heme and each of seven site-specific fluorescent probes. We have found both an equilibrium unfolding intermediate and a distinct refolding intermediate from kinetics studies. Depending on the protein region monitored, we observed either two-state or three-state denaturation transitions. The unfolding intermediate associated with three-state folding exhibited native contacts in β-sheet and C-terminal helix regions. We probed the formation of a refolding intermediate by time-resolved fluorescence energy transfer between residue 110 and the heme using a continuous flow mixer. The intermediate ensemble, a heterogeneous mixture of compact and extended polypeptides, forms in a millisecond, substantially slower than the ∼100-μs formation of a burst-phase intermediate in cytochrome c. The surprising finding is that, unlike for cytochrome c, there is an observable folding intermediate, but no microsecond burst phase in the folding kinetics of the structurally related thermostable protein.
Organizational Affiliation:
Beckman Institute, California Institute of Technology, Pasadena, CA 91125, USA.