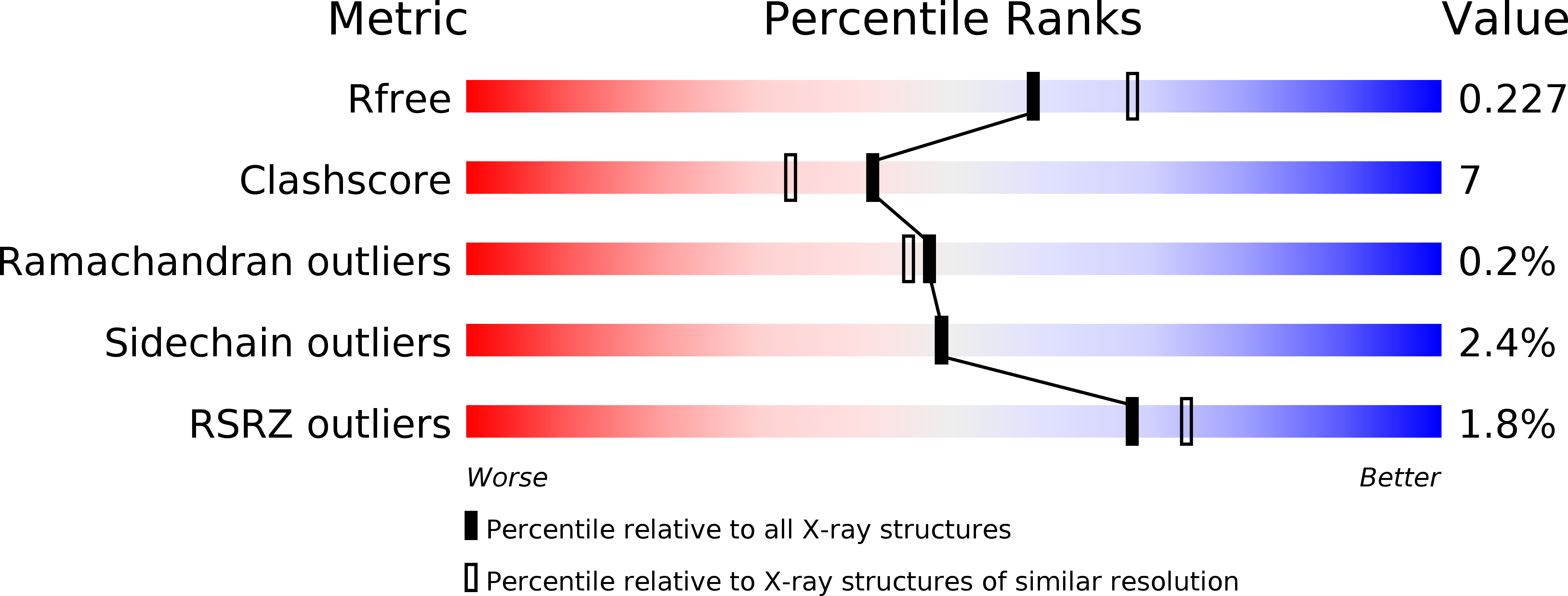
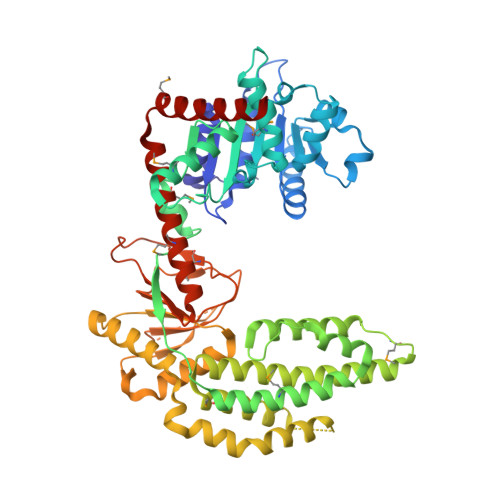
The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Civril, F., Bennett, M., Moldt, M., Deimling, T., Witte, G., Schiesser, S., Carell, T., Hopfner, K.P.(2011) EMBO Rep 12: 1127-1134
- PubMed: 21979817
- DOI: https://doi.org/10.1038/embor.2011.190
- Primary Citation of Related Structures:
3TBK - PubMed Abstract:
RIG-I detects cytosolic viral dsRNA with 5' triphosphates (5'-ppp-dsRNA), thereby initiating an antiviral innate immune response. Here we report the crystal structure of superfamily 2 (SF2) ATPase domain of RIG-I in complex with a nucleotide analogue. RIG-I SF2 comprises two RecA-like domains 1A and 2A and a helical insertion domain 2B, which together form a 'C'-shaped structure. Domains 1A and 2A are maintained in a 'signal-off' state with an inactive ATP hydrolysis site by an intriguing helical arm. By mutational analysis, we show surface motifs that are critical for dsRNA-stimulated ATPase activity, indicating that dsRNA induces a structural movement that brings domains 1A and 2A/B together to form an active ATPase site. The structure also indicates that the regulatory domain is close to the end of the helical arm, where it is well positioned to recruit 5'-ppp-dsRNA to the SF2 domain. Overall, our results indicate that the activation of RIG-I occurs through an RNA- and ATP-driven structural switch in the SF2 domain.
Organizational Affiliation:
Department of Biochemistry at the Gene Center, Ludwig-Maximilians-University Munich, Feodor-Lynen-Strasse 25, Munich 81377, Germany.