Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones
Cai, X., Wang, R., Filloux, A., Waksman, G., Meng, G.(2011) PLoS One 6: e16583-e16583
- PubMed: 21304995
- DOI: https://doi.org/10.1371/journal.pone.0016583
- Primary Citation of Related Structures:
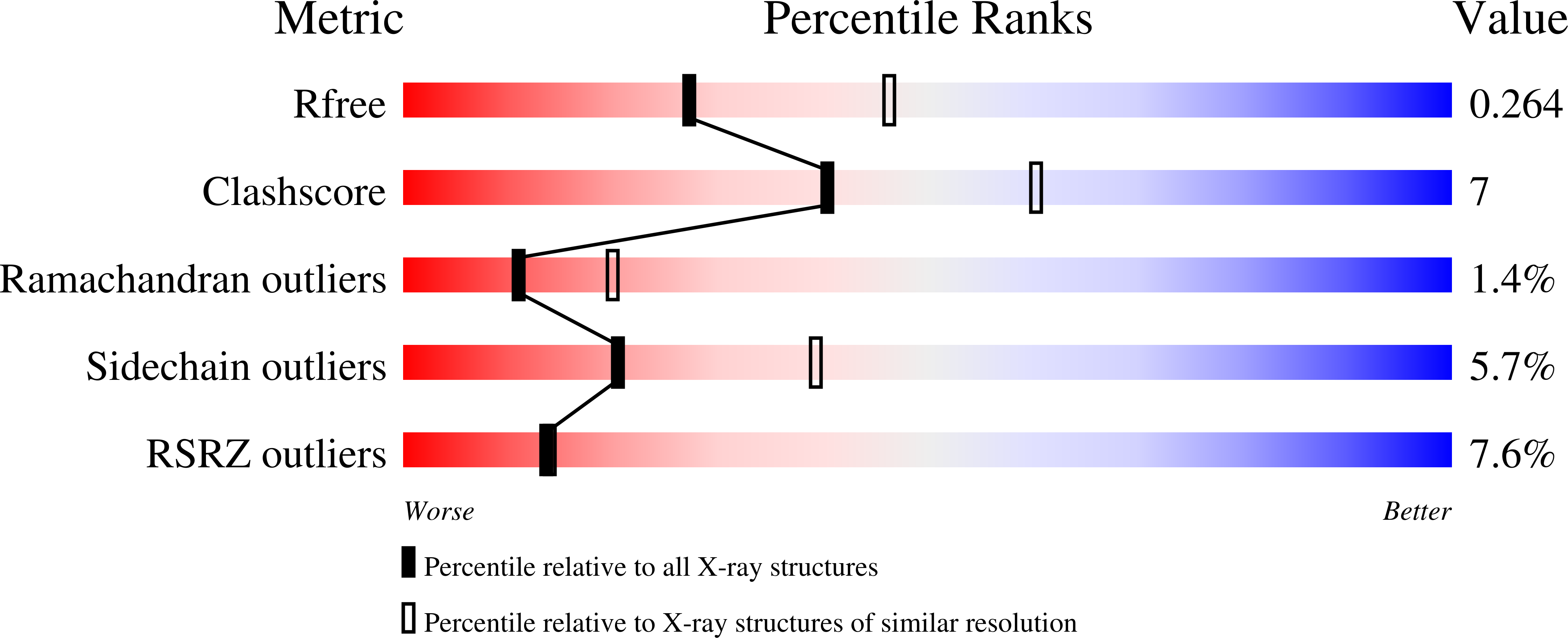
3Q48 - PubMed Abstract:
Pseudomonas aeruginosa, an important human pathogen, is estimated to be responsible for ∼10% of nosocomial infections worldwide. The pathogenesis of P. aeruginosa starts from its colonization in the damaged tissue or medical devices (e.g. catheters, prothesis and implanted heart valve etc.) facilitated by several extracellular adhesive factors including fimbrial pili. Several clusters containing fimbrial genes have been previously identified on the P. aeruginosa chromosome and named cup[1]. The assembly of the CupB pili is thought to be coordinated by two chaperones, CupB2 and CupB4. However, due to the lack of structural and biochemical data, their chaperone activities remain speculative. In this study, we report the 2.5 Å crystal structure of P. aeruginosa CupB2. Based on the structure, we further tested the binding specificity of CupB2 and CupB4 towards CupB1 (the presumed major pilus subunit) and CupB6 (the putative adhesin) using limited trypsin digestion and strep-tactin pull-down assay. The structural and biochemical data suggest that CupB2 and CupB4 might play different, but not redundant, roles in CupB secretion. CupB2 is likely to be the chaperone of CupB1, and CupB4 could be the chaperone of CupB4:CupB5:CupB6, in which the interaction of CupB4 and CupB6 might be mediated via CupB5.
Organizational Affiliation:
State Key Laboratory of Medical Genomics, Shanghai Institute of Hematology, Rui-Jin Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, People's Republic of China.