LARP4B binds to the PABC1 MLLE domain via a variant PAM2 motif
Grimm, C., Pelz, J.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polyadenylate-binding protein 1 | 82 | Homo sapiens | Mutation(s): 0 Gene Names: PABPC1, PAB1, PABP1, PABPC2 | 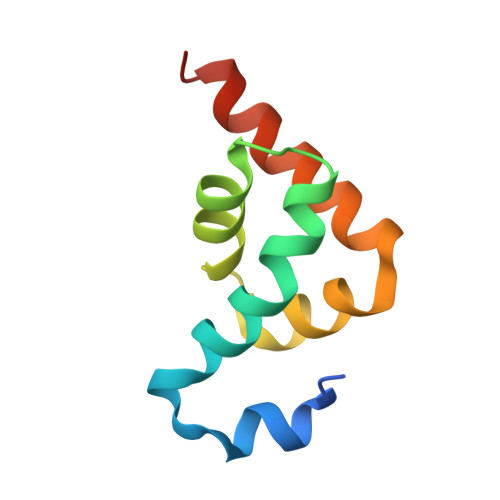 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P11940 (Homo sapiens) Explore P11940 Go to UniProtKB: P11940 | |||||
PHAROS: P11940 GTEx: ENSG00000070756 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P11940 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| La-related protein 4B | 15 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q92615 (Homo sapiens) Explore Q92615 Go to UniProtKB: Q92615 | |||||
PHAROS: Q92615 GTEx: ENSG00000107929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92615 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 29.15 | α = 90 |
| b = 57.194 | β = 90 |
| c = 59.514 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DNA | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |