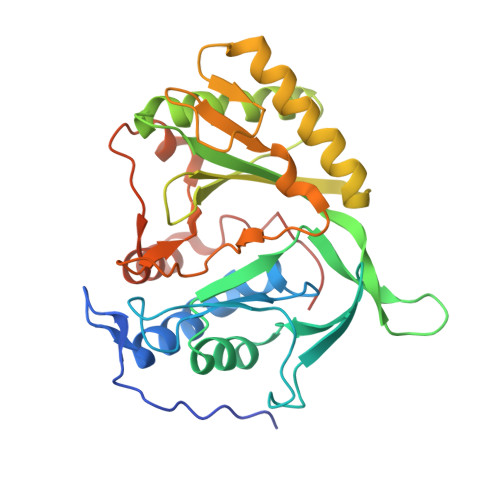
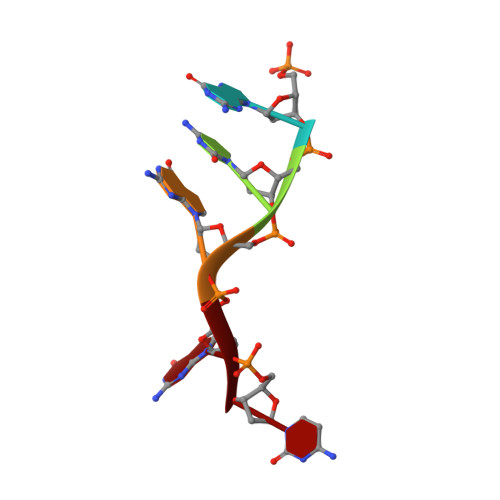
Structure of a Preternary Complex Involving a Prokaryotic NHEJ DNA Polymerase.
Brissett, N.C., Martin, M.J., Pitcher, R.S., Bianchi, J., Juarez, R., Green, A.J., Fox, G.C., Blanco, L., Doherty, A.J.(2011) Mol Cell 41: 221-231
- PubMed: 21255731
- DOI: https://doi.org/10.1016/j.molcel.2010.12.026
- Primary Citation of Related Structures:
3PKY - PubMed Abstract:
In many prokaryotes, a specific DNA primase/polymerase (PolDom) is required for nonhomologous end joining (NHEJ) repair of DNA double-strand breaks (DSBs). Here, we report the crystal structure of a catalytically active conformation of Mycobacterium tuberculosis PolDom, consisting of a polymerase bound to a DNA end with a 3' overhang, two metal ions, and an incoming nucleotide but, significantly, lacking a primer strand. This structure represents a polymerase:DNA complex in a preternary intermediate state. This polymerase complex occurs in solution, stabilizing the enzyme on DNA ends and promoting nucleotide extension of short incoming termini. We also demonstrate that the invariant Arg(220), contained in a conserved loop (loop 2), plays an essential role in catalysis by regulating binding of a second metal ion in the active site. We propose that this NHEJ intermediate facilitates extension reactions involving critically short or noncomplementary DNA ends, thus promoting break repair and minimizing sequence loss during DSB repair.
Organizational Affiliation:
Genome Damage and Stability Centre, University of Sussex, Brighton BN1 9RQ, UK.