The structural basis for membrane binding and pore formation by lymphocyte perforin.
Law, R.H., Lukoyanova, N., Voskoboinik, I., Caradoc-Davies, T.T., Baran, K., Dunstone, M.A., D'Angelo, M.E., Orlova, E.V., Coulibaly, F., Verschoor, S., Browne, K.A., Ciccone, A., Kuiper, M.J., Bird, P.I., Trapani, J.A., Saibil, H.R., Whisstock, J.C.(2010) Nature 468: 447-451
- PubMed: 21037563
- DOI: https://doi.org/10.1038/nature09518
- Primary Citation of Related Structures:
3NSJ - PubMed Abstract:
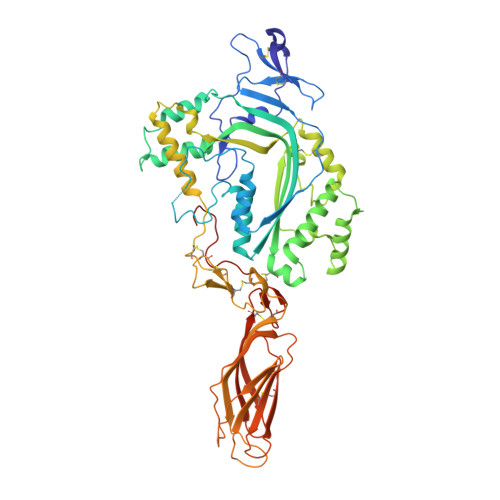
Natural killer cells and cytotoxic T lymphocytes accomplish the critically important function of killing virus-infected and neoplastic cells. They do this by releasing the pore-forming protein perforin and granzyme proteases from cytoplasmic granules into the cleft formed between the abutting killer and target cell membranes. Perforin, a 67-kilodalton multidomain protein, oligomerizes to form pores that deliver the pro-apoptopic granzymes into the cytosol of the target cell. The importance of perforin is highlighted by the fatal consequences of congenital perforin deficiency, with more than 50 different perforin mutations linked to familial haemophagocytic lymphohistiocytosis (type 2 FHL). Here we elucidate the mechanism of perforin pore formation by determining the X-ray crystal structure of monomeric murine perforin, together with a cryo-electron microscopy reconstruction of the entire perforin pore. Perforin is a thin 'key-shaped' molecule, comprising an amino-terminal membrane attack complex perforin-like (MACPF)/cholesterol dependent cytolysin (CDC) domain followed by an epidermal growth factor (EGF) domain that, together with the extreme carboxy-terminal sequence, forms a central shelf-like structure. A C-terminal C2 domain mediates initial, Ca(2+)-dependent membrane binding. Most unexpectedly, however, electron microscopy reveals that the orientation of the perforin MACPF domain in the pore is inside-out relative to the subunit arrangement in CDCs. These data reveal remarkable flexibility in the mechanism of action of the conserved MACPF/CDC fold and provide new insights into how related immune defence molecules such as complement proteins assemble into pores.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Monash University, Clayton, Melbourne, Victoria 3800, Australia.