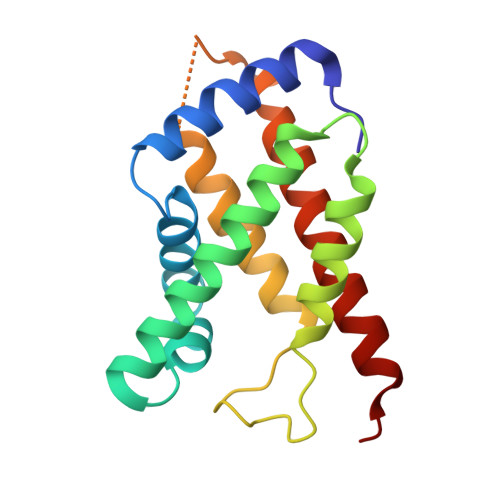
Structure and properties of a bis-histidyl ligated globin from Caenorhabditis elegans.
Yoon, J., Herzik, M.A., Winter, M.B., Tran, R., Olea, C., Marletta, M.A.(2010) Biochemistry 49: 5662-5670
- PubMed: 20518498
- DOI: https://doi.org/10.1021/bi100710a
- Primary Citation of Related Structures:
3MVC - PubMed Abstract:
Globins are heme-containing proteins that are best known for their roles in oxygen (O(2)) transport and storage. However, more diverse roles of globins in biology are being revealed, including gas and redox sensing. In the nematode Caenorhabditis elegans, 33 globin or globin-like genes were recently identified, some of which are known to be expressed in the sensory neurons of the worm and linked to O(2) sensing behavior. Here, we describe GLB-6, a novel globin-like protein expressed in the neurons of C. elegans. Recombinantly expressed full-length GLB-6 contains a heme site with spectral features that are similar to those of other bis-histidyl ligated globins, such as neuroglobin and cytoglobin. In contrast to these globins, however, ligands such as CO, NO, and CN(-) do not bind to the heme in GLB-6, demonstrating that the endogenous histidine ligands are likely very tightly coordinated. Additionally, GLB-6 exhibits rapid two-state autoxidation kinetics in the presence of physiological O(2) levels as well as a low redox potential (-193 +/- 2 mV). A high-resolution (1.40 A) crystal structure of the ferric form of the heme domain of GLB-6 confirms both the putative globin fold and bis-histidyl ligation and also demonstrates key structural features that can be correlated with the unusual ligand binding and redox properties exhibited by the full-length protein. Taken together, the biochemical properties of GLB-6 suggest that this neural protein would most likely serve as a physiological sensor for O(2) in C. elegans via redox signaling and/or electron transfer.
Organizational Affiliation:
California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA.