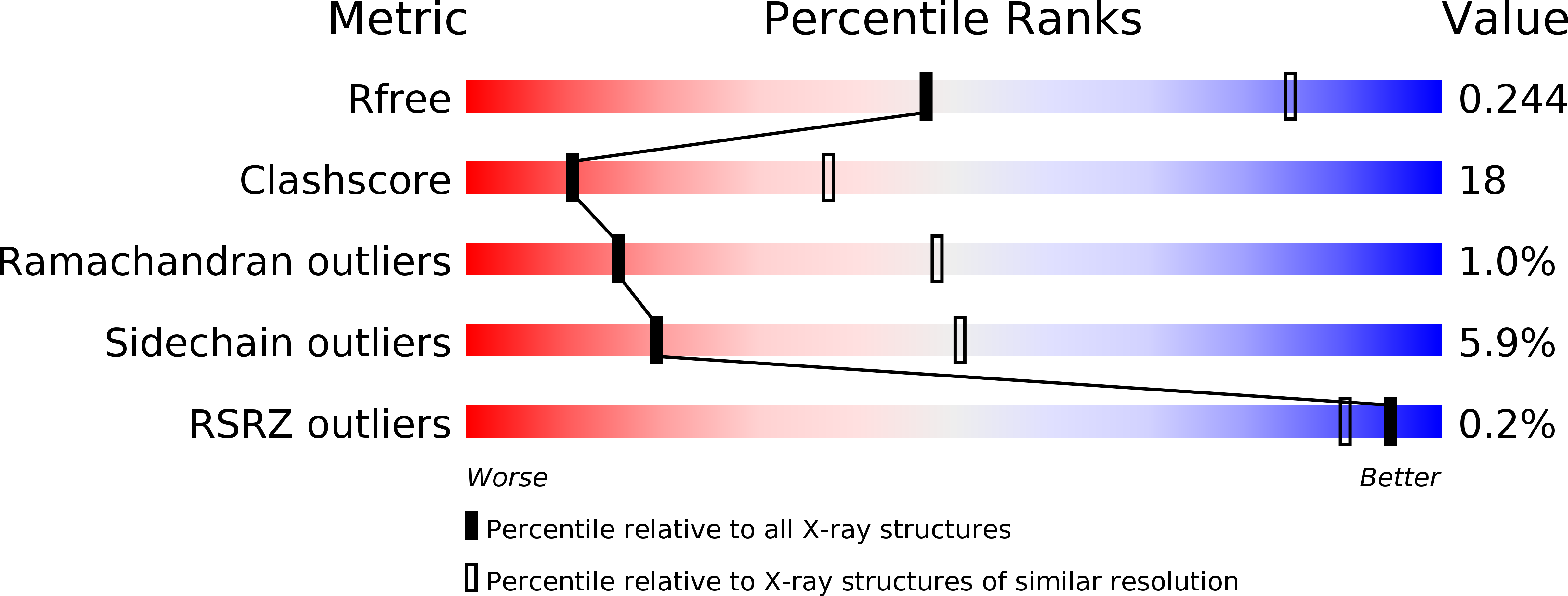
Monovalent interactions of galectin-1.
Salomonsson, E., Larumbe, A., Tejler, J., Tullberg, E., Rydberg, H., Sundin, A., Khabut, A., Frejd, T., Lobsanov, Y.D., Rini, J.M., Nilsson, U.J., Leffler, H.(2010) Biochemistry 49: 9518-9532
- PubMed: 20873803
- DOI: https://doi.org/10.1021/bi1009584
- Primary Citation of Related Structures:
3M2M - PubMed Abstract:
Galectin-1, a β-galactoside binding lectin involved in immunoregulation and cancer, binds natural and many synthetic multivalent glycoconjugates with an apparent glycoside cluster effect, that is, affinity above and beyond what would be expected from the concentration of the determinant sugar. Here we have analyzed the mechanism of such cluster effects in solution at physiological concentration using a fluorescence anisotropy assay with a novel fluorescent high-affinity galectin-1 binding probe. The interaction of native dimeric and monomeric mutants of rat and human galectin-1 with mono- and divalent small molecules, fetuin, asialofetuin, and human serum glycoproteins was analyzed. Surprisingly, high-affinity binding did not depend much on the dimeric state of galectin-1 and thus is due mainly to monomeric interactions of a single carbohydrate recognition domain. The mechanism for this is unknown, but one possibility includes additional interactions that high-affinity ligands make with an extended binding site on the carbohydrate recognition domain. It follows that such weak additional interactions must be important for the biological function of galectin-1 and also for the design of galectin-1 inhibitors.
Organizational Affiliation:
Institute of Laboratory Medicine, Lund University, Lund, Sweden.