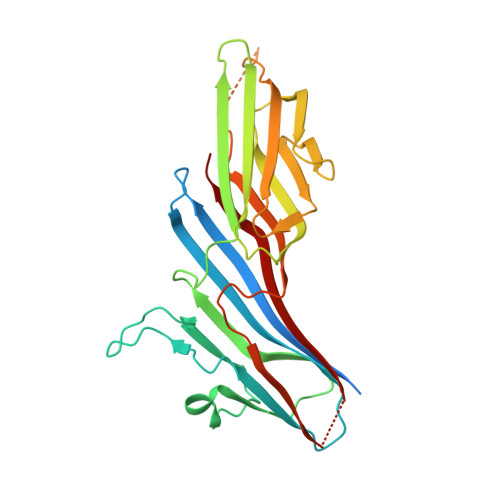
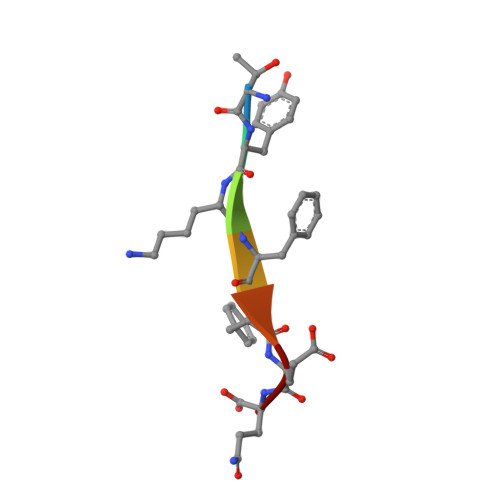
Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Burgos, P.V., Mardones, G.A., Rojas, A.L., daSilva, L.L., Prabhu, Y., Hurley, J.H., Bonifacino, J.S.(2010) Dev Cell 18: 425-436
- PubMed: 20230749
- DOI: https://doi.org/10.1016/j.devcel.2010.01.015
- Primary Citation of Related Structures:
3L81 - PubMed Abstract:
Adaptor protein 4 (AP-4) is the most recently discovered and least well-characterized member of the family of heterotetrameric adaptor protein (AP) complexes that mediate sorting of transmembrane cargo in post-Golgi compartments. Herein, we report the interaction of an YKFFE sequence from the cytosolic tail of the Alzheimer's disease amyloid precursor protein (APP) with the mu4 subunit of AP-4. Biochemical and X-ray crystallographic analyses reveal that the properties of the APP sequence and the location of the binding site on mu4 are distinct from those of other signal-adaptor interactions. Disruption of the APP-AP-4 interaction decreases localization of APP to endosomes and enhances gamma-secretase-catalyzed cleavage of APP to the pathogenic amyloid-beta peptide. These findings demonstrate that APP and AP-4 engage in a distinct type of signal-adaptor interaction that mediates transport of APP from the trans-Golgi network (TGN) to endosomes, thereby reducing amyloidogenic processing of the protein.
Organizational Affiliation:
Cell Biology and Metabolism Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.