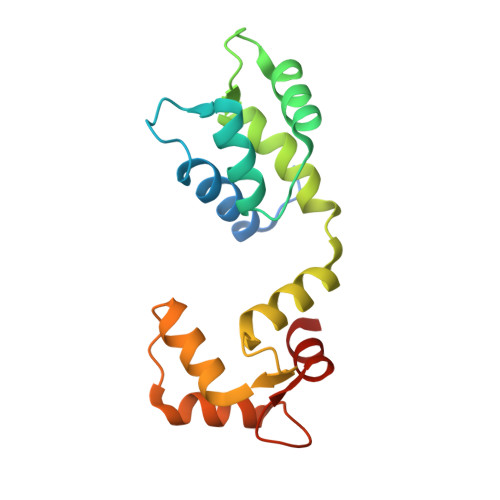
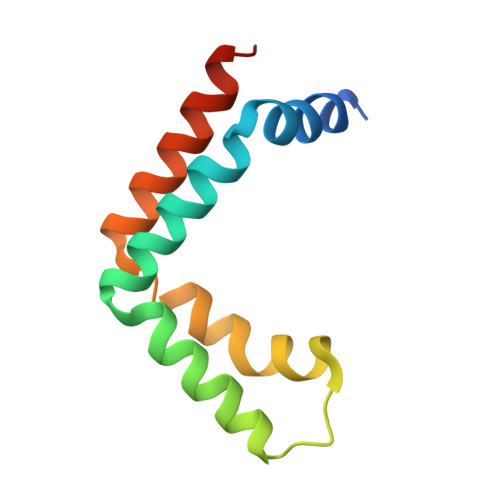
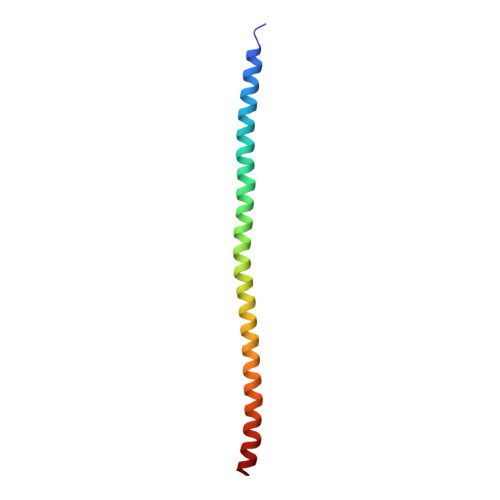Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Jani, D., Lutz, S., Marshall, N.J., Fischer, T., Kohler, A., Ellisdon, A.M., Hurt, E., Stewart, M.(2009) Mol Cell 33: 727-737
- PubMed: 19328066
- DOI: https://doi.org/10.1016/j.molcel.2009.01.033
- Primary Citation of Related Structures:
3FWB, 3FWC - PubMed Abstract:
The yeast Sac3:Cdc31:Sus1:Thp1 (TREX-2) complex facilitates the repositioning and association of actively transcribing genes with nuclear pores (NPCs)-"gene gating"-that is central to integrating transcription, processing, and mRNA nuclear export. We present here the crystal structure of Sus1 and Cdc31 bound to a central region of Sac3 (the CID domain) that is crucial for its function. Sac3(CID) forms a long, gently undulating alpha helix around which one Cdc31 and two Sus1 chains are wrapped. Sus1 has an articulated helical hairpin fold that facilitates its wrapping around Sac3. In vivo studies using engineered mutations that selectively disrupted binding of individual chains to Sac3 indicated that Sus1 and Cdc31 function synergistically to promote NPC association of TREX-2 and mRNA nuclear export. These data indicate Sac3(CID) provides a scaffold within TREX-2 to integrate interactions between protein complexes to facilitate the coupling of transcription and mRNA export during gene expression.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.