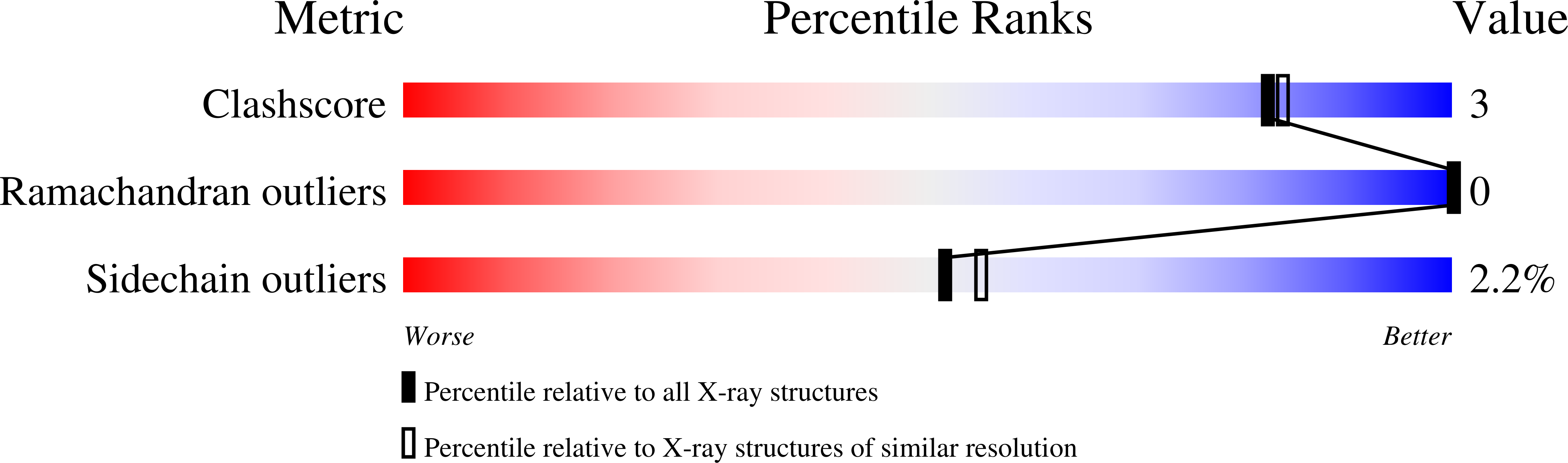A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Iwai, W., Yamada, T., Kurihara, K., Ohnishi, Y., Kobayashi, Y., Tanaka, I., Takahashi, H., Kuroki, R., Tamada, T., Niimura, N.(2009) Acta Crystallogr D Biol Crystallogr 65: 1042-1050
- PubMed: 19770501
- DOI: https://doi.org/10.1107/S090744490902770X
- Primary Citation of Related Structures:
3FHP - PubMed Abstract:
Neutron diffraction data for T(6) porcine insulin were collected to 2.1 A resolution from a single crystal partly deuterated by exchange of mother liquor. A maximum-likelihood structure refinement was undertaken using the neutron data and the structure was refined to a residual of 0.179. The hydrogen-bonding network of the central core of the hexamer was observed and the charge balance between positively charged Zn ions and their surrounding structure was interpreted by considering the protonation and/or deprotonation states and interactions of HisB10, water and GluB13. The observed double conformation of GluB13 was essential to interpreting the charge balance and could be compared with the structure of a dried crystal of T(6) human insulin at 100 K. Differences in the dynamic behaviour of the water molecules coordinating the upper and lower Zn ions were observed and interpreted. The hydrogen bonds in the insulin molecules, as well as those involving HisB10 and GluB13, are discussed. The hydrogen/deuterium (H/D) exchange ratios of the amide H atoms of T(6) porcine insulin in crystals were obtained and showed that regions highly protected from H/D exchange are concentrated in the centre of a helical region of the B chains. From the viewpoint of soaking time versus H/D-exchange ratios, the amide H atoms can be classified into three categories.
Organizational Affiliation:
Graduate School of Science and Engineering, Ibaraki University, Hitachi, Ibaraki 316-8511, Japan.