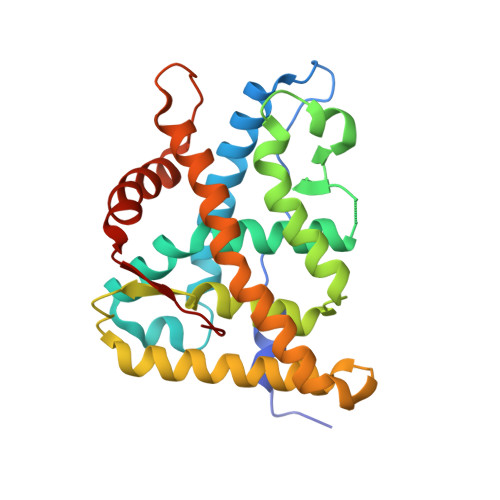
The first X-ray crystal structure of the glucocorticoid receptor bound to a non-steroidal agonist.
Madauss, K.P., Bledsoe, R.K., Mclay, I., Stewart, E.L., Uings, I.J., Weingarten, G., Williams, S.P.(2008) Bioorg Med Chem Lett 18: 6097-6099
- PubMed: 18952422
- DOI: https://doi.org/10.1016/j.bmcl.2008.10.021
- Primary Citation of Related Structures:
3E7C - PubMed Abstract:
The amino-pyrazole 2,6-dichloro-N-ethyl benzamide 1 is a selective GR agonist with dexamethasone-like in vitro potency. Its X-ray crystal structure in the GR LBD (Glucocorticoid ligand-binding domain) is described and compared to other reported structures of steroidal GR agonists in the GR LBD (3E7C).
Organizational Affiliation:
Department of Computational and Structural Chemistry, Molecular Discovery Research, GlaxoSmithKline, 5 Moore Drive, Research Triangle Park, NC 27709, USA. kevin.p.madauss@gsk.com