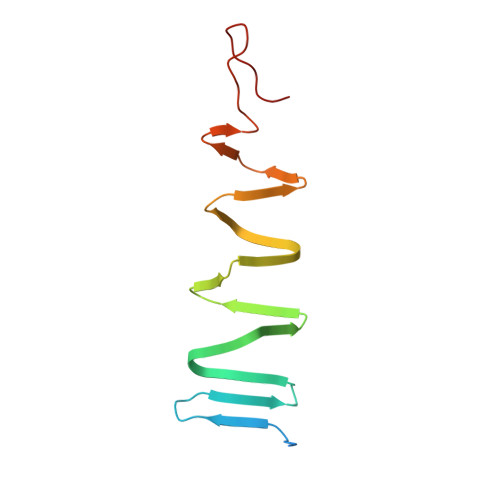
The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure
Yamashita, E., Nakagawa, A., Takahashi, J., Tsunoda, K., Yamada, S., Takeda, S.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 837-841
- PubMed: 21821878
- DOI: https://doi.org/10.1107/S1744309111005999
- Primary Citation of Related Structures:
3AQJ - PubMed Abstract:
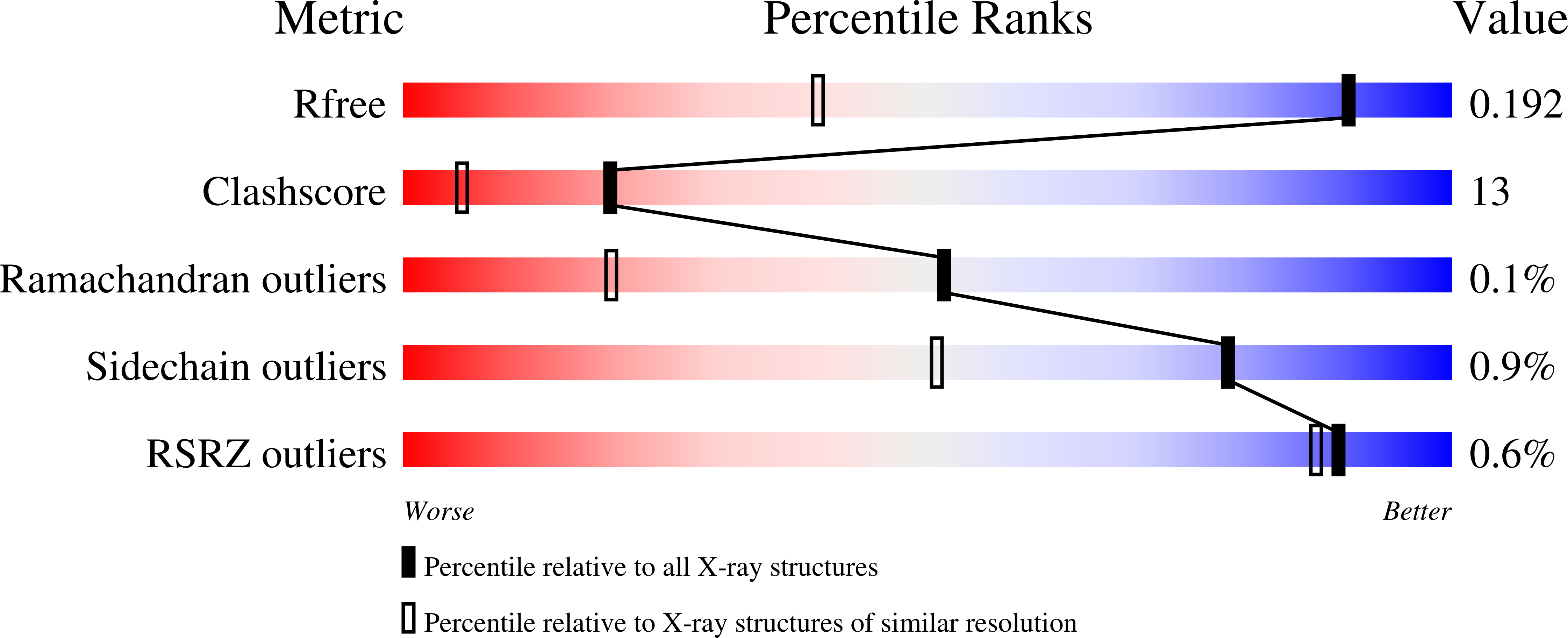
The adsorption and infection of bacteriophage P2 is mediated by tail fibres and tail spikes. The tail spikes on the tail baseplate are used to irreversibly adsorb to the host cells. Recently, a P2 phage tail-spike protein, gpV, was purified and it was shown that a C-terminal domain, Ser87-Leu211, is sufficient for the binding of gpV to host Escherichia coli membranes [Kageyama et al. (2009), Biochemistry, 48, 10129-10135]. In this paper, the crystal structure of the C-terminal domain of P2 gpV is reported. The structure is a triangular pyramid and looks like a spearhead composed of an intertwined β-sheet, a triple β-helix and a metal-binding region containing iron, calcium and chloride ions.
Organizational Affiliation:
Institute for Protein Research, Osaka University, Suita, Osaka, Japan.