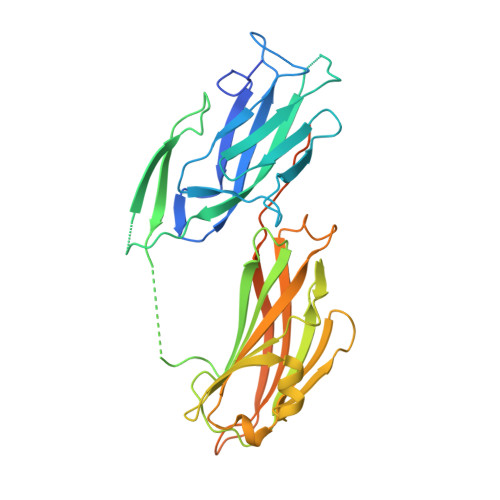
The Enterococcus faecalis MSCRAMM ACE binds its ligand by the Collagen Hug model
Liu, Q., Ponnuraj, K., Xu, Y., Ganesh, V.K., Sillanpaa, J., Murray, B.E., Narayana, S.V.L., Hook, M.(2007) J Biol Chem 282: 19629-19637
- PubMed: 17392280
- DOI: https://doi.org/10.1074/jbc.M611137200
- Primary Citation of Related Structures:
2Z1P - PubMed Abstract:
We have determined the crystal structure of the ligand binding segment of the Enterococcus faecalis collagen binding MSCRAMM ACE (microbial surface components recognizing adhesive matrix molecules adhesin of collagen from enterococci). This segment is composed of two subdomains, N(1) and N(2), each adopting an IgG-like fold and forming a putative collagen binding surface at the interface between the two subdomains. This structure is very similar to that recently reported for CNA, the collagen binding MSCRAMM of Staphylococcus aureus, for which a unique ligand binding mechanism called the Collagen Hug was proposed. We suggest that ACE binds collagen by a similar mechanism and present the first biochemical evidence for this binding model. Replacing residues in the putative collagen binding trench of ACE N(2) with Ala residues affected collagen binding. A closed conformation of ACE stabilized by an engineered disulfide bond is unable to bind collagen. Finally, the importance of the residues in the N(2) extension in stabilizing the MSCRAMM-ligand complex is demonstrated by selected point and truncation mutations.
Organizational Affiliation:
Center for Extracellular Matrix Biology, Texas A&M University System Health Science Center, Albert B. Alkek Institute of Biosciences and Technology, Houston, Texas 77030, USA.