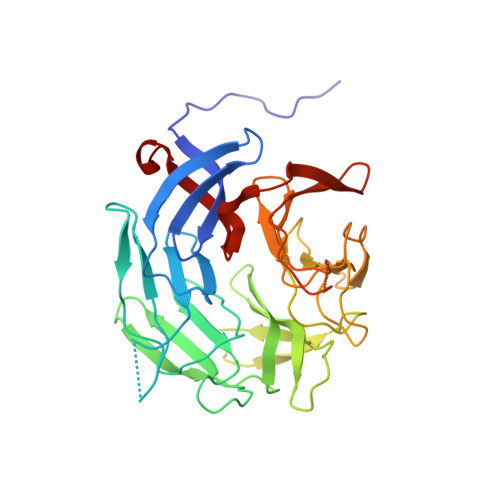
Novel beta-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension.
Gray, C.H., McGarry, L.C., Spence, H.J., Riboldi-Tunnicliffe, A., Ozanne, B.W.(2009) J Biol Chem 284: 30498-30507
- PubMed: 19726686
- DOI: https://doi.org/10.1074/jbc.M109.023259
- Primary Citation of Related Structures:
2WOZ - PubMed Abstract:
Kelch-related protein 1 (Krp1) is up-regulated in oncogene-transformed fibroblasts. The Kelch repeats interact directly with the actin-binding protein Lasp-1 in membrane ruffles at the tips of pseudopodia, where both proteins are necessary for pseudopodial elongation. Herein, we investigate the molecular basis for this interaction. Probing an array of overlapping decapeptides of Rattus norvegicus (Rat) Krp1 with recombinant Lasp-1 revealed two binding sites; one ((317)YDPMENECYLT(327)) precedes the first of five Kelch repeats, and the other ((563)TEVNDIWKYEDD(574)) is in the last of the five Kelch repeats. Mutational analysis established that both binding sites are necessary for Krp1-Lasp-1 interaction in vitro and function in vivo. The crystal structure of the C-terminal domain of rat Krp1 (amino acids 289-606) reveals that both binding sites are brought into close proximity by the formation of a novel six-bladed beta-propeller, where the first blade is not formed by a Kelch repeat.
Organizational Affiliation:
Invasion and Metastasis Laboratory, Beatson Institute for Cancer Research, Garscube Estate, Glasgow G61 1BD, Scotland, United Kingdom. c.gray@beatson.gla.ac.uk