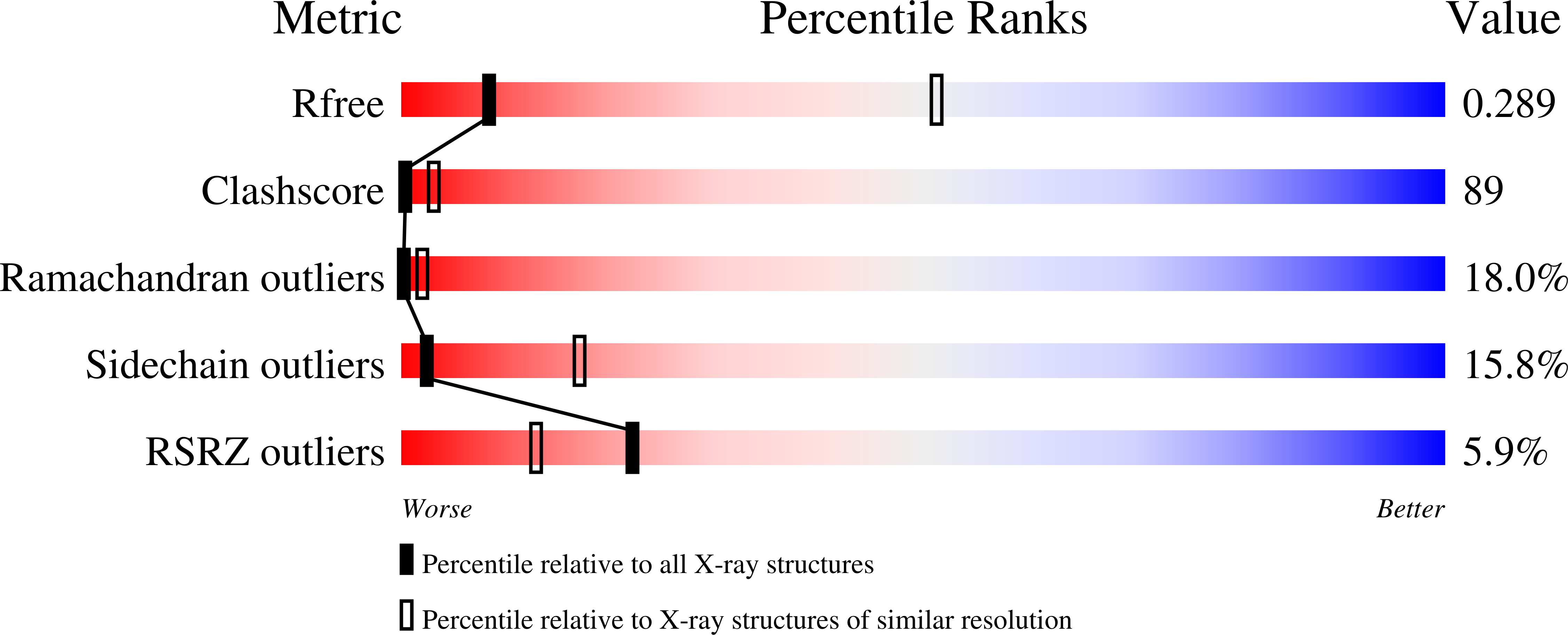
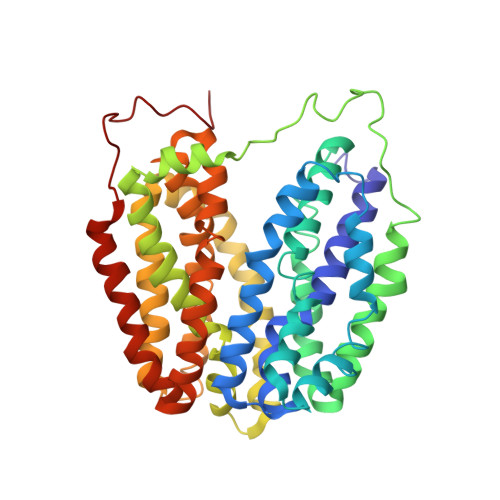
Structural Determination of Wild-Type Lactose Permease.
Guan, L., Mirza, O., Verner, G., Iwata, S., Kaback, H.R.(2007) Proc Natl Acad Sci U S A 104: 15294
- PubMed: 17881559
- DOI: https://doi.org/10.1073/pnas.0707688104
- Primary Citation of Related Structures:
2V8N - PubMed Abstract:
Here we describe an x-ray structure of wild-type lactose permease (LacY) from Escherichia coli determined by manipulating phospholipid content during crystallization. The structure exhibits the same global fold as the previous x-ray structures of a mutant that binds sugar but cannot catalyze translocation across the membrane. LacY is organized into two six-helix bundles with twofold pseudosymmetry separated by a large interior hydrophilic cavity open only to the cytoplasmic side and containing the side chains important for sugar and H(+) binding. To initiate transport, binding of sugar and/or an H(+) electrochemical gradient increases the probability of opening on the periplasmic side. Because the inward-facing conformation represents the lowest free-energy state, the rate-limiting step for transport may be the conformational change leading to the outward-facing conformation.
Organizational Affiliation:
Department of Physiology, University of California, Los Angeles, CA 90095-1662, USA.