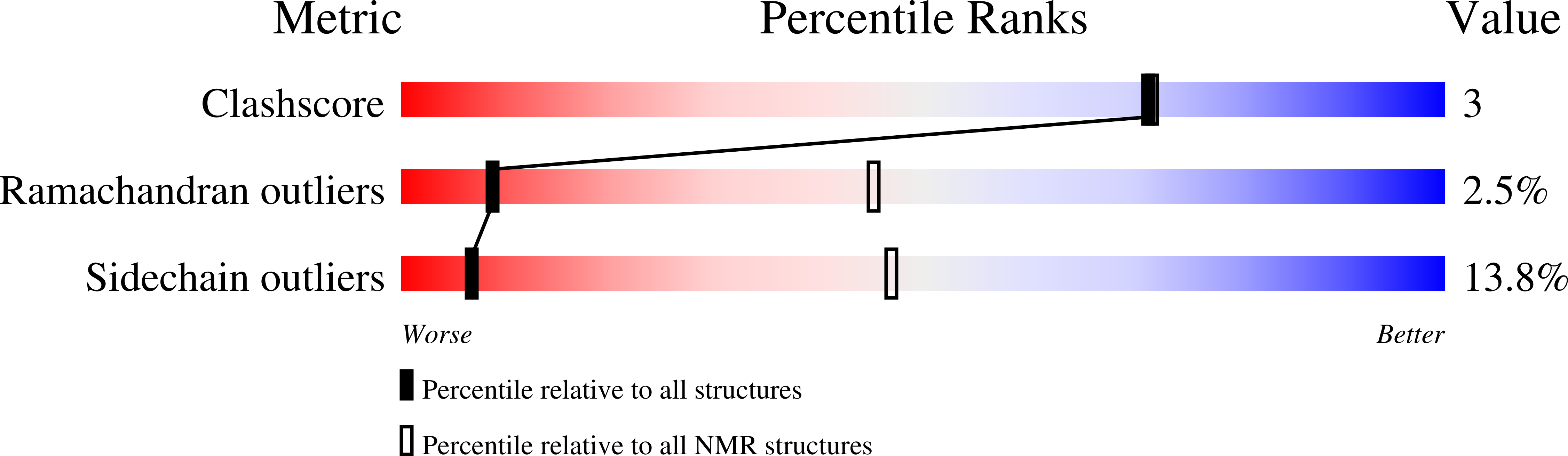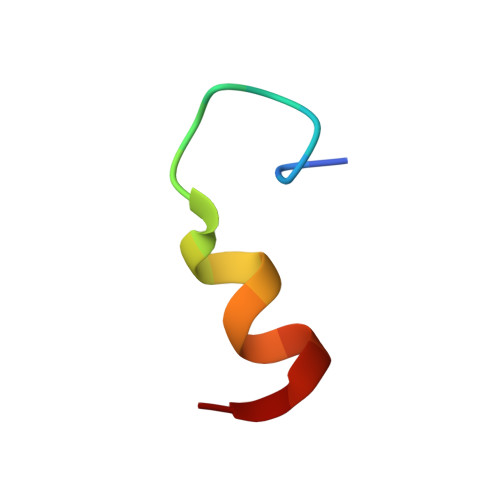Solution Structure of LC4 Transmembrane Segment of CCR5
Miyamoto, K., Togiya, K.(2011) PLoS One 6: e20452-e20452
- PubMed: 21647380
- DOI: https://doi.org/10.1371/journal.pone.0020452
- Primary Citation of Related Structures:
2RRS - PubMed Abstract:
CC-chemokine receptor 5 (CCR5) is a specific co-receptor allowing the entry of human immunodeficiency virus type 1 (HIV-1). The LC4 region in CCR5 is required for HIV-1 entry into the cells. In this study, the solution structure of LC4 in SDS micelles was elucidated by using standard 1H two-dimensional NMR spectroscopy, circular dichroism, and fluorescence quenching. The LC4 structure adopts two helical structures, whereas the C-terminal part remains unstructured. The positions in which LC4 binds to the HIV-1 inhibitory peptide LC5 were determined by docking calculations in addition to NMR data. The poses showed the importance of the hydrophobic interface of the assembled structures. The solution structure of LC4 elucidated in the present work provides a structural basis for further studies on the HIV-1 inhibitory function of the LC4 region.
Organizational Affiliation:
Department of Pharmaceutical Health Care, Faculty of Pharmaceutical Sciences, Himeji Dokkyo University, Himeji, Hyogo, Japan. miyamoto@himeji-du.ac.jp