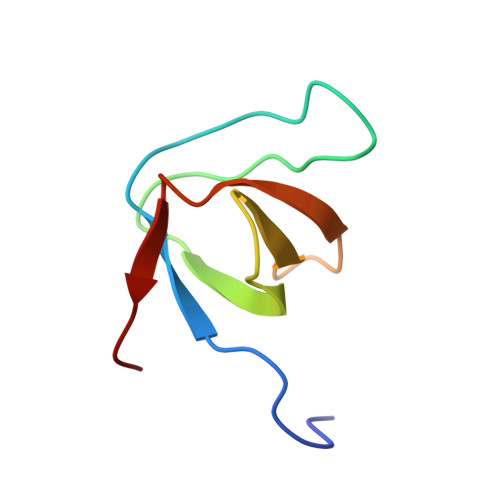
Structural basis of PxxDY motif recognition in SH3 binding
Aitio, O., Hellman, M., Kesti, T., Kleino, I., Samuilova, O., Paakkonen, K., Tossavainen, H., Saksela, K., Permi, P.(2008) J Mol Biol 382: 167-178
- PubMed: 18644376
- DOI: https://doi.org/10.1016/j.jmb.2008.07.008
- Primary Citation of Related Structures:
2K2M, 2ROL - PubMed Abstract:
We have determined the solution structure of epidermal growth factor receptor pathway substrate 8 (Eps8) L1 Src homology 3 (SH3) domain in complex with the PPVPNPDYEPIR peptide from the CD3epsilon cytoplasmic tail. Our structure reveals the distinct structural features that account for the unusual specificity of the Eps8 family SH3 domains for ligands containing a PxxDY motif instead of canonical PxxP ligands. The CD3epsilon peptide binds Eps8L1 SH3 in a class II orientation, but neither adopts a polyproline II helical conformation nor engages the first proline-binding pocket of the SH3 ligand binding interface. Ile531 of Eps8L1 SH3, instead of Tyr or Phe residues typically found in this position in SH3 domains, renders this hydrophobic pocket smaller and nonoptimal for binding to conventional PxxP peptides. A positively charged arginine at position 512 in the n-Src loop of Eps8L1 SH3 plays a key role in PxxDY motif recognition by forming a salt bridge to D7 of the CD3epsilon peptide. In addition, our structural model suggests a hydrogen bond between the hydroxyl group of the aromatic ring of Y8 and the carboxyl group of E496, thus explaining the critical role of the PxxDY motif tyrosine residue in binding to Eps8 family SH3. These finding have direct implications also for understanding the atypical binding specificity of the amino-terminal SH3 of the Nck family proteins.
Organizational Affiliation:
Program in Structural Biology and Biophysics, Institute of Biotechnology/NMR Laboratory, University of Helsinki, FI-00014 Helsinki, Finland.