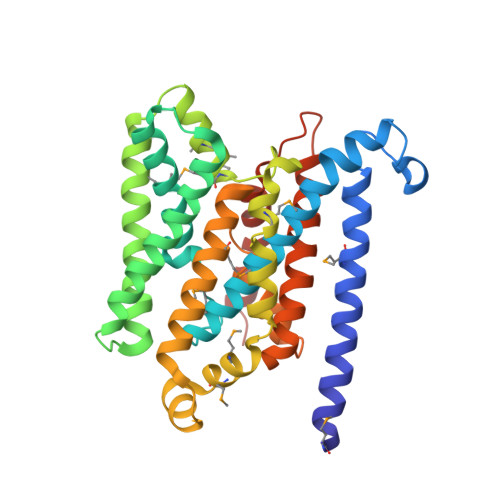
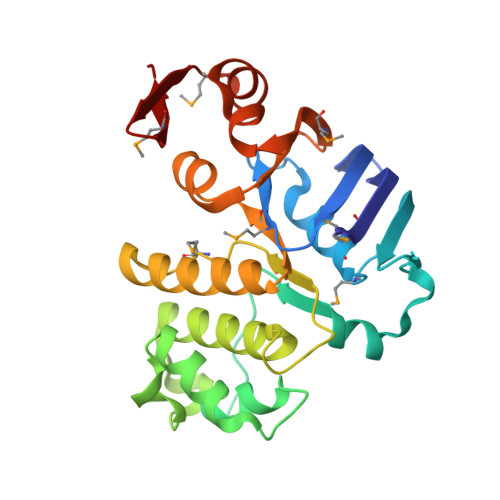
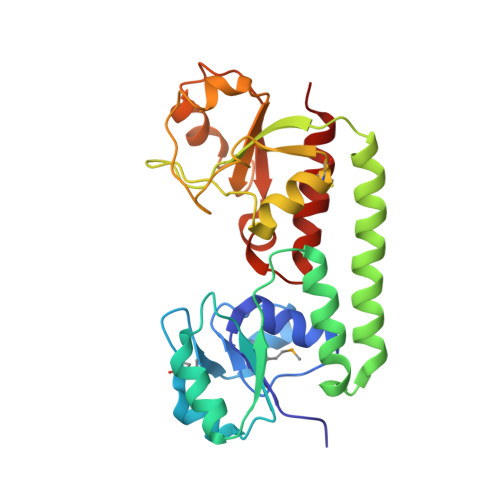
Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF.
Hvorup, R.N., Goetz, B.A., Niederer, M., Hollenstein, K., Perozo, E., Locher, K.P.(2007) Science 317: 1387-1390
- PubMed: 17673622
- DOI: https://doi.org/10.1126/science.1145950
- Primary Citation of Related Structures:
2QI9 - PubMed Abstract:
BtuCD is an adenosine triphosphate-binding cassette (ABC) transporter that translocates vitamin B12 from the periplasmic binding protein BtuF into the cytoplasm of Escherichia coli. The 2.6 angstrom crystal structure of a complex BtuCD-F reveals substantial conformational changes as compared with the previously reported structures of BtuCD and BtuF. The lobes of BtuF are spread apart, and B12 is displaced from the binding pocket. The transmembrane BtuC subunits reveal two distinct conformations, and the translocation pathway is closed to both sides of the membrane. Electron paramagnetic resonance spectra of spin-labeled cysteine mutants reconstituted in proteoliposomes are consistent with the conformation of BtuCD-F that was observed in the crystal structure. A comparison with BtuCD and the homologous HI1470/71 protein suggests that the structure of BtuCD-F may reflect a posttranslocation intermediate.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zurich, HPK D14.3, 8093 Zurich, Switzerland.