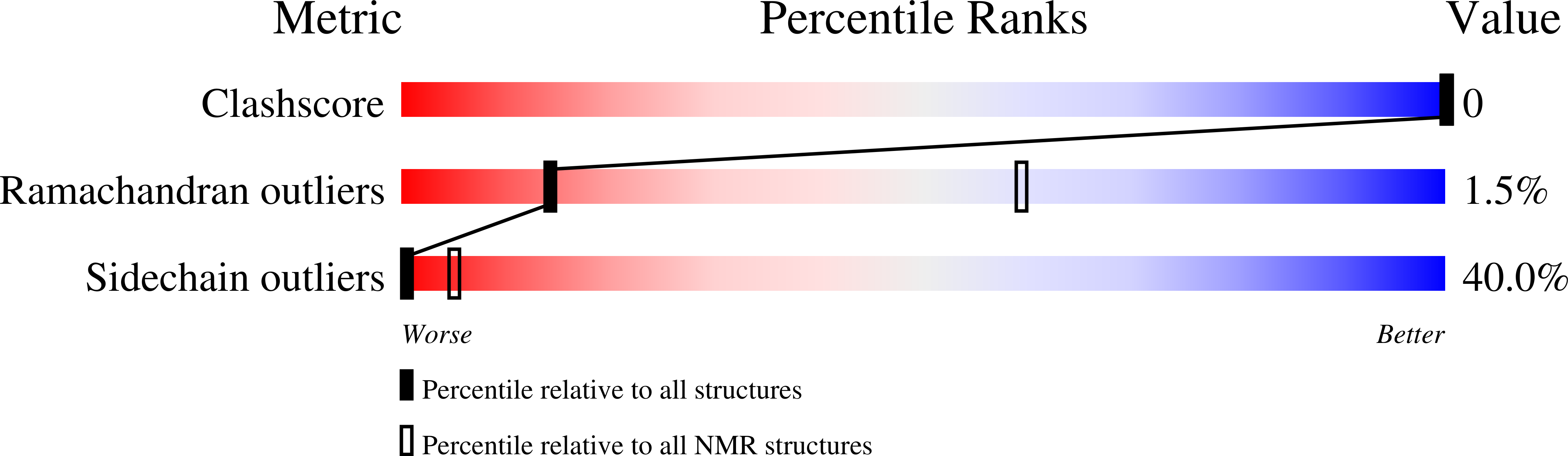Evolutionary Pareto-optimization of stably folding peptides
Gronwald, W., Hohm, T., Hoffmann, D.(2008) BMC Bioinformatics 9: 109-109
- PubMed: 18284690
- DOI: https://doi.org/10.1186/1471-2105-9-109
- Primary Citation of Related Structures:
2PPZ - PubMed Abstract:
As a rule, peptides are more flexible and unstructured than proteins with their substantial stabilizing hydrophobic cores. Nevertheless, a few stably folding peptides have been discovered. This raises the question whether there may be more such peptides that are unknown as yet. These molecules could be helpful in basic research and medicine. As a method to explore the space of conformationally stable peptides, we have developed an evolutionary algorithm that allows optimization of sequences with respect to several criteria simultaneously, for instance stability, accessibility of arbitrary parts of the peptide, etc. In a proof-of-concept experiment we have perturbed the sequence of the peptide Villin Headpiece, known to be stable in vitro. Starting from the perturbed sequence we applied our algorithm to optimize peptide stability and accessibility of a loop. Unexpectedly, two clusters of sequences were generated in this way that, according to our criteria, should form structures with higher stability than the wild-type. The structures in one of the clusters possess a fold that markedly differs from the native fold of Villin Headpiece. One of the mutants predicted to be stable was selected for synthesis, its molecular 3D-structure was characterized by nuclear magnetic resonance spectroscopy, and its stability was measured by circular dichroism. Predicted structure and stability were in good agreement with experiment. Eight other sequences and structures, including five with a non-native fold are provided as bona fide predictions. The results suggest that much more conformationally stable peptides may exist than are known so far, and that small fold classes could comprise well-separated sub-folds.
Organizational Affiliation:
Institute for Functional Genomics, University of Regensburg, Josef-Engert-Strasse 9, 93053 Regensburg, Germany. wolfram.gronwald@klinik.uni-regensburg.de