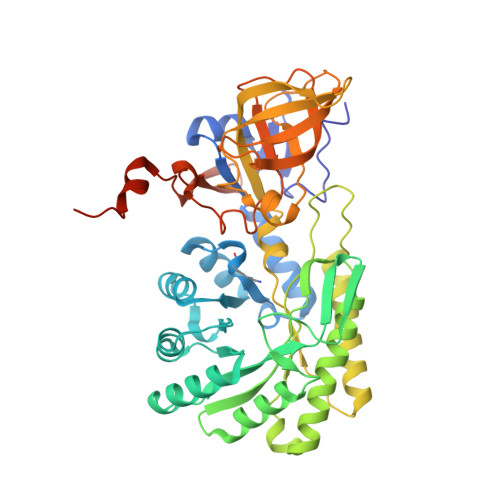
The three-dimensional structure of diaminopimelate decarboxylase from Mycobacterium tuberculosis reveals a tetrameric enzyme organisation.
Weyand, S., Kefala, G., Svergun, D.I., Weiss, M.S.(2009) J Struct Funct Genomics 10: 209-217
- PubMed: 19543810
- DOI: https://doi.org/10.1007/s10969-009-9065-z
- Primary Citation of Related Structures:
2O0T - PubMed Abstract:
The three-dimensional structure of the enzyme diaminopimelate decarboxylase from Mycobacterium tuberculosis has been determined in a new crystal form and refined to a resolution of 2.33 A. The monoclinic crystals contain one tetramer exhibiting D(2)-symmetry in the asymmetric unit. The tetramer exhibits a donut-like structure with a hollow interior. All four active sites are accessible only from the interior of the tetrameric assembly. Small-angle X-ray scattering indicates that in solution the predominant oligomeric species of the protein is a dimer, but also that higher oligomers exist at higher protein concentrations. The observed scattering data are best explained by assuming a dimer-tetramer equilibrium with about 7% tetramers present in solution. Consequently, at the elevated protein concentrations in the crowded environment inside the cell the observed tetramer may constitute the biologically relevant functional unit of the enzyme.
Organizational Affiliation:
EMBL Hamburg Outstation, c/o DESY, Hamburg, Germany.